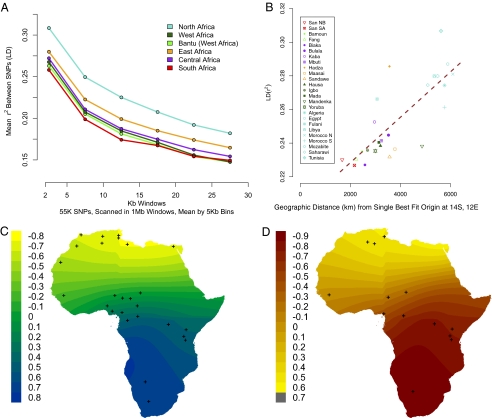
Fig. 2.
Genome-wide LD and Fst in African populations. (A) Each line represents the LD decay averaged across populations within each of six geographic regions; regions are described in SI Appendix, Table S1. LD (r2) between SNPs was calculated in sliding 1-Mb windows. The r2 estimates were binned by the genetic distance between SNPs in 5-Kb bins. HG populations have the lowest LD curves (SI Appendix, Fig. S13 shows population-specific LD decay curves). (B) We assessed a possible point of origin by regressing LD onto geographic distance. Regression for the single best fit for geographic origin is shown, with a correlation coefficient of 0.78, to the point 14°S, 12°E. (C) Map is shown using mean LD within 0–5 Kb. The highest correlation coefficient in blue indicates the best fit with a potential geographic origin. Crosses indicate the geographic coordinates of the sampled populations. (D) We assessed a possible point of origin by regressing Fst between African populations and Europe onto geographic distance, using a waypoint in the Near East. Populations were grouped for Fst analysis; crosses indicate the geographic mean for each population grouping.