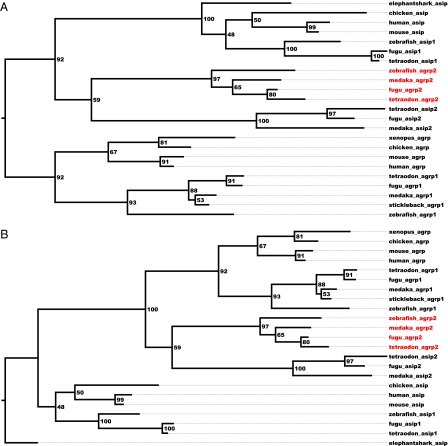
Fig. 1.
Phylogeny of the agouti gene family members. The full-length sequences from Braasch and Postlethwait (1) were aligned (except sequences where the reference was to unspecified Contig regions) using MAFFT-EINSI 6.624b with default settings and 10 rebuilds. RAxML-III was used with the fast and easy settings: bootstrap maximum likelihood, Whelan and Goldman (WAG) model, estimate proportion of invariable sites, empirical base frequencies, and 100 bootstraps using easyrax.pl as interface. (A) This dendrogram is rooted on the agrp clusters. (B) This dendrogram is equivalent to A but rerooted on the elephant shark full-length sequence (Callorhinchus milii). The difference is that now the A2 cluster, containing the agrp2 and asip2 sequences, is grouped with the agrp cluster instead of the asip cluster. Bipartitions trees were viewed in Fig Tree 1.3.1 (http://tree.bio.ed.ac.uk/software/figtree/).