Abstract
Molecular, cellular, and animal-based studies have recently exposed circadian clocks as critical regulators of energy balance. Invariably, mouse models of genetically manipulated circadian clock components display features indicative of altered lipid/fatty acid metabolism, including differential adiposity and circulating lipids. The purpose of this minireview is to provide a comprehensive summary of current knowledge regarding the regulation of fatty acid metabolism by distinct cell autonomous circadian clocks. The implications of these recent findings for cardiometabolic disease and human health are discussed.
Keywords: Energy Metabolism, Fatty Acid Metabolism, Fatty Acid Oxidation, Transcription Regulation, Triglyceride, Circadian Clock
Introduction
Ranging from the transcriptome to whole body energy balance, considerable evidence exists in support of the concept that fatty acid (FA)2 metabolism is regulated in a time of day-dependent fashion in mammals, including humans. That whole organism FA metabolism is influenced by daily fluctuations in activity/nutritional status is widely recognized (e.g. increased FA oxidation rates during periods of intermittent fasting). Manipulation of dietary macronutrient quantity and quality, as well as the timing of meals throughout the 24-h day, markedly impacts whole body FA metabolism. Time of day-dependent oscillations in whole body FA metabolism occur concomitantly with daily changes in circulating metabolites (e.g. non-esterified FAs, triglycerides) and various neurohumoral factors known to influence triglyceride turnover in a significant manner (e.g. insulin, norepinephrine). Collectively, these observations have led to suggestions that fluctuations in external/environmental influences (e.g. light and food availability) primarily drive daily rhythms in FA metabolism via associated oscillations in circulating nutrients and metabolically relevant neural and hormonal inputs. However, recent molecular studies present compelling evidence that cell autonomous circadian clocks act as molecular cornerstones in metabolic homeostasis and energy balance.
Circadian Clocks Influence Whole Body Energy Balance
Within the last decade, a wealth of information has firmly established the mammalian circadian clock as an important regulator of energy homeostasis (1–4). In turn, metabolism has emerged as an integral clock component, such that perturbations in the metabolic milieu influence the clock mechanism (5). The mammalian circadian clock is a cell autonomous, transcriptionally based mechanism composed of an expanding list of core proteins that generate a series of feedback loops, resulting in rhythmic expression of clock components and downstream target genes (6, 7). Great effort has been made to identify clock output genes (8). Both hypothesis-generating and hypothesis-testing methodologies have been utilized to investigate time of day-dependent (diurnal and circadian) transcriptional oscillations in tissues/cells from wild-type and genetically modified rodent models (9–17). The reported observations unequivocally demonstrate that a large proportion of transcripts oscillating in a 24-h manner encode critical regulators of multiple energy metabolism pathways. These transcriptional oscillations likely translate to cell autonomous circadian clock-mediated daily fluctuations in energy metabolism/balance (5).
Multiple mouse models of genetically manipulated circadian clock components display gross phenotypic alterations indicative of changes in energy balance (e.g. body weight and adiposity). Genetic manipulation of CLOCK and BMAL1, two transcription factors that reside within the core of the mammalian mechanism, provides prime examples (18, 19). Turek and co-workers (20) reported that ClockΔ19 mutant mice (harboring a dominant-negative mutation in the clock gene) exhibit a metabolic syndrome-like phenotype (including increased adiposity and dyslipidemia) on a C57BL/6J background. In marked contrast, genetic ablation of BMAL1, the heterodimerization partner of CLOCK, results in leanness (21). Somewhat surprisingly, both ClockΔ19 and Bmal1 null mice exhibit increased insulin tolerance (relative to wild-type controls) (22). However, glucose intolerance is observed in both models, likely consequent to impaired insulin secretion (23–25). Indeed, persistence of circadian oscillations in insulin secretion by cultured βTC-3 insulinoma cells is indicative of direct clock control (26). Underscoring the impact of circadian clocks on energy balance, ClockΔ19 mutant mice show a modest increase in food intake concomitant with decreased energy expenditure, resulting in positive energy balance (consistent with the reported obesity propensity) (20).
In terms of metabolic phenotyping, ClockΔ19 mutant and Bmal1 null mice are among the most extensively characterized models of genetic perturbation of the mammalian circadian clock. It is noteworthy that several additional models of clock disruption have also been characterized metabolically, albeit to varying extents. These include genetic manipulation of the cryptochrome, period, Rev-erbα, and retinoid-related orphan receptor-α genes (27–31). Furthermore, putative clock-controlled components hypothesized to act as key molecular links between the circadian clock and energy balance have been investigated (e.g. AMP-activated protein kinase, GSK3β, nocturin, and PGC1α/β) (32–36). In all cases, genetic manipulation of clock components or putative clock outputs results in distinct perturbations in energy homeostasis. Despite this wealth of recent molecular knowledge, the current understanding of the mechanisms by which cell autonomous clocks directly influence flux through discrete metabolic pathways is less clear. Energy balance is the product of the complex interaction between genes (affording predisposition toward leanness or obesity) and environment (inclusive of nutrition, physical activity, and the neurohumoral milieu). Circadian clocks may potentially impact, or be impacted by, any of these factors. For example, diurnal rhythms in food intake and physical activity are attenuated in ClockΔ19 mutant mice, which is associated with loss of rhythms in various humoral factors (20). Because of the ubiquitous nature of the circadian clock (present within virtually all mammalian cells), the host of processes that this timekeeper likely influences (estimated to regulate >10% of the transcriptome) and the fact that distinct clock components may exert differing effects, defining the mechanism(s) by which a cell autonomous clock directly regulates distinct metabolic processes such as FA metabolism, present a significant challenge.
Direct Regulation of FA Metabolism by Cell Autonomous Clocks?
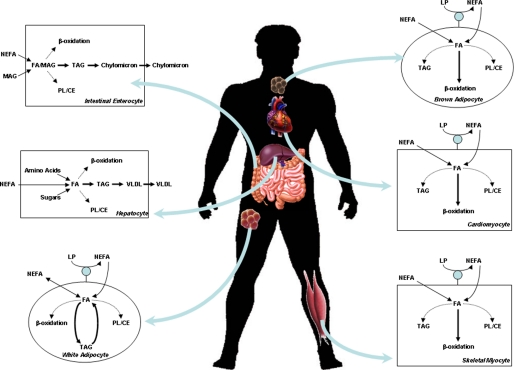
FA metabolism encompasses multiple processes (including dietary lipid digestion and absorption, lipoprotein metabolism, de novo FA synthesis, triglyceride turnover, phospholipid metabolism, and FA oxidation) orchestrated by distinct cell types (including enterocytes, hepatocytes, adipocytes, and myocytes) (Fig. 1). In each case, evidence exists supporting the concept that circadian clocks influence these processes either directly or indirectly. Classically, indirect evidence of regulation by circadian clocks has included persistence of a 24-h rhythmic pattern under constant environmental conditions, as well as loss of this rhythmicity in models of genetic disruption of circadian clock components (e.g. ClockΔ19 mutant and Bmal1 null mice) (6). To unveil metabolic processes directly regulated by a cell autonomous clock, 24-h rhythmic patterns must persist in cultured cells, and/or rhythmicity should be lost in models of cell type-specific clock disruption. The most robust conclusions related to metabolism are based on measurement of metabolic fluxes as opposed to indirect markers (e.g. gene and protein expression or activity of metabolic enzymes, steady-state levels of metabolites in plasma or tissues). For the sake of brevity, this minireview will focus on pathways unique to FA metabolism. As such, little attention will be given to de novo FA synthesis due to unavoidable overlap with carbohydrate and amino acid metabolism.
FIGURE 1.
FA metabolism within metabolically relevant tissues. NEFA, non-esterified FA; MAG, monoacylglycerol; TAG, triacylglycerol; PL, phospholipid; CE, cholesterol ester; LP, lipoprotein; VLDL, very low density lipoprotein.
Dietary Lipid Digestion/Absorption
Multiple aspects of digestion/absorption exhibit a time-of-day dependence in mammals, including food intake, gastric emptying, and intestinal motility, as well as the expression and activity of critical digestive enzymes and nutrient transport systems (37, 38). This is exemplified by lipid digestive capacity and absorption. Various enzymes/proteins involved in these processes exhibit diurnal variations at gene, protein, and activity levels, as well as flux (39, 40). Lipid absorption has been shown to peak during the beginning of the active/awake period, a time at which food intake often increases (40). This peak in lipid absorption is associated with greater activity of digestive lipases, as well as the enzymes involved in chylomicron synthesis within intestinal enterocytes (40, 41). Pan and Hussain (40) have shown that time of day-dependent oscillations in these parameters are attenuated in ClockΔ19 mutant mice on the C57BL/6J background, resulting in increased capacity for lipid absorption. In contrast, the ClockΔ19 mutation on the ICR background impairs lipid digestion/absorption (42). In addition, disruption of nocturin, a clock output gene encoding a deadenylase, confers resistance to high fat feeding-induced obesity, a phenotype that has been attributed to impaired lipid digestion/absorption (34).
An important question relates to whether cell autonomous circadian clocks, perhaps within intestinal enterocytes, directly regulate lipid digestion/absorption. Microsomal triglyceride transfer protein (MTP) is a critical enzyme in lipoprotein (e.g. chylomicron) synthesis, and levels of MTP correlate closely with plasma lipids and lipoproteins (39). MTP activity oscillates in a time of day-dependent manner in enterocytes, peaking at the beginning of the active period (40). Importantly, oscillations persist in cultured Huh-7 cells and are abolished when the circadian clock is genetically disrupted (43). Collectively, these studies have led to the hypothesis that direct control of MTP (and/or other relevant enzymes/proteins) by the enterocyte circadian clock may play an important role in time of day-dependent oscillations in lipid digestion/absorption.
Lipoprotein Metabolism
Plasma total triglyceride and lipoproteins exhibit time of day-dependent oscillations, as do de novo lipogenesis and cholesterol synthesis (43–46). Oscillations in circulating triglyceride/lipoproteins cannot be accounted for solely by rhythms in food intake, as they persist during prolonged fasting (47). Furthermore, these oscillations are attenuated in ClockΔ19 mice, suggesting mediation by the circadian clock in some manner (43). Similar to intestinal enterocytes, MTP expression (gene and protein) oscillates in a time of day-dependent manner in wild-type (but not ClockΔ19 mutant) livers (43). Rhythms in lipoprotein-associated triglyceride hydrolysis could potentially contribute toward oscillations in circulating triglycerides. Lipoprotein lipase (LPL) activity demonstrates distinct time of day-dependent oscillations in peripheral tissues, which are essentially antiphase in muscle compared with adipose tissue (48). LPL activity appears to be under direct circadian clock control (49). This has led Gimble and Floyd (49) to hypothesize that cell autonomous circadian clocks may channel lipoprotein-derived triglyceride utilization into adipose tissue and muscle at distinct times of the day. Collectively, these observations suggest that cell autonomous circadian clocks may drive both lipoprotein synthesis (i.e. intestinal enterocyte and hepatocyte clocks) and utilization (i.e. myocyte and adipocyte clocks).
Triglyceride Turnover
Given that ubiquitous genetic manipulation of clock components influences adiposity, several laboratories have attempted to establish links between circadian clocks with adipocyte function. Consistent with the lean phenotype observed in Bmal1 null mice, studies by Shimba et al. (50) defined a critical role for Bmal1 in adipogenesis. Although direct measures of metabolic flux were not reported in these studies (i.e. lipolysis/lipogenesis), the investigators observed that Bmal1 directly influences the activity of transcription factors known to modulate expression of numerous triglyceride metabolism enzymes (e.g. peroxisome proliferator-activated receptor-γ (PPARγ), CCAAT/enhancer-binding protein-α/β, etc.) (50). Subsequently, BMAL1 has been shown to associate directly with PPARγ (51). Similar to Bmal1 null mice, PER2-deficient mice exhibit a lean phenotype, and molecular studies reveal a direct PER2-PPARγ interaction (29). More recently, we have generated a novel mouse model wherein the circadian clock is disrupted in an adipocyte-specific manner in vivo, through targeted expression of the CLOCKΔ19 mutant protein in adipocytes (termed adipocyte-specific clock mutant (ACM) mice) (52). Similar to the ubiquitous ClockΔ19 mutant mouse, ACM mice exhibit an obesity phenotype, suggesting that the adipocyte clock directly influences adiposity (20, 52). Microarray studies on white adipose tissue from wild-type versus ACM mice revealed differential gene expression of multiple triglyceride metabolism enzymes (e.g. FA transporter, glycerol kinase) (52). However, to date, triglyceride turnover rates have not been assessed in adipose tissue of this or other genetic models of clock disruption.
A significant question remaining is whether cell autonomous clocks directly regulate triglyceride turnover (at a metabolic flux level). This question was recently addressed using the heart as a model of an insulin-sensitive, metabolically active organ. Like adipose tissue, the heart actively synthesizes and degrades triglyceride in a time of day-dependent manner, and abnormalities in myocardial triglyceride turnover have been linked to cardiovascular disease progression (53–56). Adipose triglyceride lipase expression in the heart is second only to that in adipose tissue, and genetic ablation of this lipase results in severe cardiac steatosis and sudden cardiac death (57). Studies using a mouse model of cardiomyocyte-specific clock mutation (CCM) revealed that this clock directly regulates triglyceride turnover, independent of extrinsic influences (54). More specifically, this cell autonomous clock promotes increased rates of lipolysis during the less active/sleep phase (54). Clock-driven attenuation of lipolysis during the active phase in turn augments triglyceride accumulation at this time (54).
Phospholipid Metabolism
Phospholipids have essential structural and signaling functions (58–61). Steady-state levels of phospholipids, rates of their synthesis, and activity of key phospholipid metabolism enzymes have been shown to exhibit diurnal variations in multiple tissues, which generally peak during the inactive/sleep phase (53, 62, 63). Multiple lines of evidence exist in support of the concept that time of day-dependent rhythms in phospholipid metabolism are driven by cell autonomous circadian clocks. For example, rhythms in phospholipid synthesis and the activity of phosphatidate phosphohydrolase persist in retinal cells during constant darkness (63). Through the use of radiolabeled tracers, Marquez et al. (64) reported that fibroblasts exhibit 24-h oscillations in phospholipid biosynthesis in culture. Importantly, the investigators reported that these circadian oscillations are absent in fibroblasts isolated from either Per1 null or ClockΔ19 mutant mice (64). Similar rhythmic patterns were reported for synthesis of individual phospholipids, such as phosphatidylethanolamine and phosphatidylcholine (64). Collectively, these observations provide substantial evidence that cell autonomous clocks directly regulate phospholipid synthesis in a time of day-dependent manner. Additional studies are required to reveal the relative contribution of extrinsic (i.e. neurohumoral) versus cell autonomous circadian clock influence on phospholipid synthesis rhythms in the in vivo setting.
FA Oxidation
A important question, in terms of common metabolic diseases (such as obesity, diabetes mellitus, and cardiovascular disease), is whether peripheral circadian clocks directly regulate FA β-oxidation. Whole body FA oxidative metabolism can be determined relatively easily by indirect calorimetry. If circadian clocks were to coordinately regulate FA oxidation in metabolically active tissues, then time of day-dependent oscillations in the respiratory exchange ratio (RER) should persist under constant darkness (i.e. circadian conditions) and should be absent in mouse models of clock disruption. Somewhat surprisingly, an exhaustive literature search was unable to identify previously published studies reporting RER during constant darkness (despite multiple studies reporting energy expenditure, body temperature, and physical activity under circadian conditions). Thus, we housed wild-type mice within calorimeters under constant environment-controlled conditions. As anticipated, circadian oscillations in RER were observed throughout the 4-week period of temperature-controlled constant darkness.3 A surprisingly similar lack of information has been published regarding time of day-dependent oscillations in RER for genetic mouse models of clock disruption. Again, despite multiple studies reporting energy expenditure, body temperature, and physical activity in these models, only one published study has reported RER oscillations in mice following disruption of a core circadian clock component. Vollmers et al. (27) found that diurnal variations in RER are absent in CRY1/CRY2 double knock-out mice. It is noteworthy that RER has recently been reported in PER2 null mice, although the data were presented as daily average values; no difference was observed between wild-type and PER2 null mice for 24-h averaged RER values (29).
Collectively, the aforementioned observations suggest that circadian clocks potentially influence whole body FA oxidation rates over the course of the day. The subsequent question relates to whether this regulation is direct or indirect. A number of factors influence FA oxidation, including substrate availability and energetic demand. As such, both food intake and physical activity, processes known to be under clock control, strongly impact whole body FA oxidation rates. Indeed, oscillations in these behaviors persist under circadian conditions and are altered (to differing extents) following genetic disruption of circadian clock components (6). This is exemplified in CRY1/CRY2 double knock-out mice, for which both food intake and RER oscillations are ablated (27). Importantly, reestablishment of time of day-dependent food intake oscillations in CRY1/CRY2 double knock-out mice through enforced restricted feeding completely restores oscillations in RER, suggesting that a significant proportion of the regulation of FA oxidation dictated by circadian clocks is secondary to regulation of pertinent behaviors (such as food intake) (27).
Similar to the rationale discussed for non-oxidative FA metabolism pathways, to determine whether a cell autonomous clock directly regulates FA oxidation rates requires measurement of metabolic fluxes in an isolated cell-based system and/or use of models in which the clock is disrupted in a cell type-specific manner. Of the cell type-specific models reported thus far, only one study has investigated rates of FA oxidation in a time of day-dependent manner. Neither wild-type rat nor mouse hearts exhibit diurnal variations in FA oxidation rates (15, 53). Furthermore, only slight (10%) differences in FA oxidation rates are observed between wild-type and CCM hearts, which are independent of time of day (15). As such, these data do not support the hypothesis that cell autonomous clocks directly regulate FA oxidation over the course of the day. Interestingly, expression of the CLOCKΔ19 mutant protein in skeletal muscle also results in a slight increase in FA oxidation rates in soleus muscles (although time-of-day dependence was not investigated) (52).
Does the Cell Autonomous Circadian Clock Directly Regulate FA Oxidation?
As outlined above, considerable published evidence suggests that cell autonomous circadian clocks directly regulate non-oxidative FA metabolism. What is less convincing at this time is whether the influence of clocks on FA oxidative metabolism is direct or indirect (e.g. secondary to behavioral and/or neurohumoral changes). At a transcriptional level, multiple genes known to encode key FA oxidation regulators oscillate in a time of day-dependent manner in metabolically active tissues (23, 27, 65). However, no data are currently available that suggest these transcriptional changes translate to significant oscillations in FA oxidation rates within a specific metabolically active tissue independent of feeding and/or activity status. Clearly, this could be due to lack of experimental findings at this time, as opposed to lack of phenomena. Indeed, in the heart, absence of oscillations in FA oxidation rates is not unexpected, given that rates are basally high, and increased energetic demands (during periods of increased contractile function) are generally met by alterations in carbohydrate metabolism, as opposed to FA oxidation (66). Whether FA oxidation rates oscillate in other metabolically active tissues (e.g. liver, skeletal muscle, adipose tissue) over the course of the day in a cell autonomous clock-dependent manner remains to be determined.
An additional possibility that should be considered relates to the primary function of circadian clocks: to provide the selective advantage of anticipation (7). Reliance on FAs as a fuel is elevated during periods of prolonged fasting and following a high fat meal. For the animal in the wild, prolongation of the overnight fast would occur when foraging upon waking was unsuccessful. Anticipating high circulating FAs at the beginning of the active period would be a selective advantage, allowing rapid activation of FA oxidation while the animal continues its forage for food and/or avoids predation. Evidence exists in support of a role for cell autonomous clocks in anticipation of fasting. For example, prolongation of the sleep phase fast in mice results in a rapid induction of transcripts known to promote FA oxidation in cardiac and skeletal muscles, a phenomenon that is markedly attenuated in clock-ablated CCM hearts (65, 67). Therefore, the potential role for cell autonomous clocks in the direct regulation of FA oxidation may not be apparent in the ad libitum fed laboratory rodent but can be exposed when time of day-dependent dietary challenges are imposed.
Implications for Dietary Intake and Energy Balance
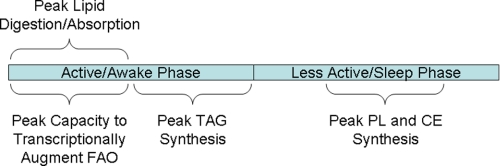
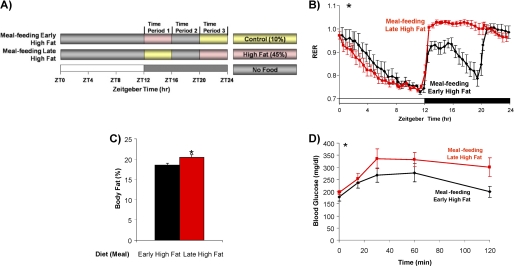
Fig. 2 summarizes current knowledge regarding temporal regulation of FA metabolism by cell autonomous circadian clocks, as highlighted above. Armed with this knowledge, one can formulate evidence-based hypotheses regarding metabolic outcomes following time of day-dependent FA challenges. In the intact animal, FA challenges are achieved easily through dietary manipulation (e.g. fasting or high fat meal feeding). Time of day-dependent high fat meal consumption may be most relevant in terms of human health in Western society. As food intake usually occurs during the active/awake phase, the metabolic consequences following high fat meal consumption at the beginning or end of this period (i.e. for breakfast or dinner, respectively) may differ dramatically. Through a series of long-term, time of day-dependent, high fat meal feeding studies, we recently examined the metabolic response to high fat feeding at different times of the day (68). Mice were provided a high fat meal either at the beginning or end of the active period. For these studies, mice consumed the same total number of daily calories, as well as the same proportion of calories from fat, carbohydrate, and protein. The only variable was the time of day at which fat (and reciprocally carbohydrate) calories were consumed. Mice fed a high fat diet during the end of the active phase were not able to activate β-oxidation and had increased body weight, increased adiposity, increased cardiac steatosis, and decreased glucose tolerance, as well as hypertriglyceridemia, hyperinsulinemia, and hyperleptinemia, compared with mice fed the same high fat meal at the beginning of the active phase (Fig. 3) (68). Thus, the time of day at which a high fat meal is consumed influences multiple cardiometabolic syndrome parameters independent of total daily caloric quantity and quality.
FIGURE 2.
Time of day-dependent oscillations in FA metabolism that are known to be influenced by circadian clocks either directly or indirectly. FAO, FA oxidation; TAG, triacylglycerol; PL, phospholipid; CE, cholesterol ester.
FIGURE 3.
Distinct high fat meal feeding regimes (A) differentially influence RER (B), adiposity (C), and glucose tolerance (D). Data were published previously in Ref. 68. *, p < 0.05 for early high fat feeding versus late high fat feeding.
The observations made in our time-of-day feeding studies are consistent with the information summarized in Fig. 2. Consumption of a high fat meal upon awakening will result in rapid digestion and absorption of dietary FAs, followed by elevation in plasma chylomicron and non-esterified FA levels. Both act as sources of FAs for metabolically active peripheral tissues (e.g. skeletal muscle, heart). Increased LPL activity in muscle (coupled with decreased activity in adipose tissue) at this time will promote channeling of lipoprotein-derived FAs to muscle (48). Transcriptional responsiveness of peripheral tissues to FAs increases at the beginning of the active phase, resulting in rapid induction of enzymes promoting FA β-oxidation, thereby increasing the capacity to utilize this carbon source for ATP generation (65). This would in turn minimize lipid accumulation (i.e. net triglyceride synthesis) in adipose and other peripheral tissues, thereby promoting leanness and insulin sensitivity. Conversely, when a high fat meal is consumed at the end of the active period (i.e. dinner), lipid digestion and absorption are likely slower at this time of the day, resulting in a prolonged elevation of plasma lipids, even into the sleep phase. Responsiveness of metabolically active peripheral tissues to FAs is not as great during the end of the active period, likely resulting in a blunted oxidation of dietary FAs. At this time, capacity for triglyceride synthesis is at a peak (54). Limited β-oxidation and increased triglyceride synthesis capacity, coupled with increased adipose tissue LPL activity, will promote adiposity. Furthermore, elevation of both plasma and intracellular lipid/FA levels during the sleep phase would provide carbon for channeling into phospholipid and cholesterol ester synthesis (53). The latter may in turn contribute to imbalanced signaling and/or lipotoxicity. Collectively, consumption of a high fat meal at the end of the active period promotes obesity and insulin resistance.
Summary
Research to date using global and tissue-specific models of circadian clock disruption provide substantial evidence that this molecular mechanism plays an important role in energy balance and lipid metabolism. Processes from digestion/absorption to cellular metabolism all appear under clock control, either direct or indirect. The knowledge gained by this research points to the importance of timing of energy intake and expenditure as critical components of energy balance. Future studies are needed to elucidate fully the roles that cell autonomous clocks play in lipid metabolism and human health.
This work was supported, in whole or in part, by National Institutes of Health Grant HL-074259 from NHLBI (to M. E. Y.). This work was also supported by Kraft Foods Inc. (to M. S. B. and M. E. Y.). This work was also supported by Kraft Foods Inc. (to M. S. B. and M. E. Y.). This minireview will be reprinted in the 2011 Minireview Compendium, which will be available in January, 2012.
M. S. Bray and M. E. Young, unpublished observations.
- FA
- fatty acid
- MTP
- microsomal triglyceride transfer protein
- LPL
- lipoprotein lipase
- PPARγ
- peroxisome proliferator-activated receptor-γ
- ACM
- adipocyte-specific clock mutant
- CCM
- cardiomyocyte-specific clock mutation
- RER
- respiratory exchange ratio.
REFERENCES
- 1. Green C. B., Takahashi J. S., Bass J. (2008) Cell 134, 728–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wijnen H., Young M. W. (2006) Annu. Rev. Genet. 40, 409–448 [DOI] [PubMed] [Google Scholar]
- 3. Rutter J., Reick M., McKnight S. L. (2002) Annu. Rev. Biochem. 71, 307–331 [DOI] [PubMed] [Google Scholar]
- 4. Sahar S., Sassone-Corsi P. (2009) Nat. Rev. Cancer 9, 886–896 [DOI] [PubMed] [Google Scholar]
- 5. Bass J., Takahashi J. S. (2010) Science 330, 1349–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Takahashi J. S., Hong H. K., Ko C. H., McDearmon E. L. (2008) Nat. Rev. Genet. 9, 764–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Edery I. (2000) Physiol. Genomics 3, 59–74 [DOI] [PubMed] [Google Scholar]
- 8. Lowrey P. L., Takahashi J. S. (2004) Annu. Rev. Genomics Hum. Genet. 5, 407–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Storch K. F., Lipan O., Leykin I., Viswanathan N., Davis F. C., Wong W. H., Weitz C. J. (2002) Nature 417, 78–83 [DOI] [PubMed] [Google Scholar]
- 10. McCarthy J. J., Andrews J. L., McDearmon E. L., Campbell K. S., Barber B. K., Miller B. H., Walker J. R., Hogenesch J. B., Takahashi J. S., Esser K. A. (2007) Physiol. Genomics 31, 86–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rudic R. D., McNamara P., Reilly D., Grosser T., Curtis A. M., Price T. S., Panda S., Hogenesch J. B., FitzGerald G. A. (2005) Circulation 112, 2716–2724 [DOI] [PubMed] [Google Scholar]
- 12. Martino T., Arab S., Straume M., Belsham D. D., Tata N., Cai F., Liu P., Trivieri M., Ralph M., Sole M. J. (2004) J. Mol. Med. 82, 256–264 [DOI] [PubMed] [Google Scholar]
- 13. Reddy A. B., Karp N. A., Maywood E. S., Sage E. A., Deery M., O'Neill J. S., Wong G. K., Chesham J., Odell M., Lilley K. S., Kyriacou C. P., Hastings M. H. (2006) Curr. Biol. 16, 1107–1115 [DOI] [PubMed] [Google Scholar]
- 14. Kornmann B., Schaad O., Bujard H., Takahashi J. S., Schibler U. (2007) PLoS Biol. 5, e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bray M. S., Shaw C. A., Moore M. W., Garcia R. A., Zanquetta M. M., Durgan D. J., Jeong W. J., Tsai J. Y., Bugger H., Zhang D., Rohrwasser A., Rennison J. H., Dyck J. R., Litwin S. E., Hardin P. E., Chow C. W., Chandler M. P., Abel E. D., Young M. E. (2008) Am. J. Physiol. Heart Circ. Physiol. 294, H1036–H1047 [DOI] [PubMed] [Google Scholar]
- 16. Yang X., Downes M., Yu R. T., Bookout A. L., He W., Straume M., Mangelsdorf D. J., Evans R. M. (2006) Cell 126, 801–810 [DOI] [PubMed] [Google Scholar]
- 17. Ptitsyn A. A., Gimble J. M. (2011) Ann. Med. 43, 1–12 [DOI] [PubMed] [Google Scholar]
- 18. Gekakis N., Staknis D., Nguyen H. B., Davis F. C., Wilsbacher L. D., King D. P., Takahashi J. S., Weitz C. J. (1998) Science 280, 1564–1569 [DOI] [PubMed] [Google Scholar]
- 19. Hogenesch J., Gu Y., Jain S., Bradfield C. (1998) Proc. Natl. Aad Sci. U.S.A. 95, 5474–5479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Turek F. W., Joshu C., Kohsaka A., Lin E., Ivanova G., McDearmon E., Laposky A., Losee-Olson S., Easton A., Jensen D. R., Eckel R. H., Takahashi J. S., Bass J. (2005) Science 308, 1043–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bunger M. K., Walisser J. A., Sullivan R., Manley P. A., Moran S. M., Kalscheur V. L., Colman R. J., Bradfield C. A. (2005) Genesis 41, 122–132 [DOI] [PubMed] [Google Scholar]
- 22. Rudic R. D., McNamara P., Curtis A. M., Boston R. C., Panda S., Hogenesch J. B., Fitzgerald G. A. (2004) PLoS Biol. 2, e377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kennaway D. J., Owens J. A., Voultsios A., Boden M. J., Varcoe T. J. (2007) Am. J. Physiol. Regul. Integr. Comp. Physiol. 293, R1528–R1537 [DOI] [PubMed] [Google Scholar]
- 24. Marcheva B., Ramsey K. M., Buhr E. D., Kobayashi Y., Su H., Ko C. H., Ivanova G., Omura C., Mo S., Vitaterna M. H., Lopez J. P., Philipson L. H., Bradfield C. A., Crosby S. D., JeBailey L., Wang X., Takahashi J. S., Bass J. (2010) Nature 466, 627–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sadacca L. A., Lamia K. A., deLemos A. S., Blum B., Weitz C. J. (2011) Diabetologia 54, 120–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Allaman-Pillet N., Roduit R., Oberson A., Abdelli S., Ruiz J., Beckmann J. S., Schorderet D. F., Bonny C. (2004) Mol. Cell. Endocrinol. 226, 59–66 [DOI] [PubMed] [Google Scholar]
- 27. Vollmers C., Gill S., DiTacchio L., Pulivarthy S. R., Le H. D., Panda S. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 21453–21458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang E. E., Liu Y., Dentin R., Pongsawakul P. Y., Liu AC., Hirota T., Nusinow D. A., Sun X., Landais S., Kodama Y., Brenner D. A., Montminy M., Kay S. A. (2010) Nat. Med. 16, 1152–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Grimaldi B., Bellet M. M., Katada S., Astarita G., Hirayama J., Amin R. H., Granneman J. G., Piomelli D., Leff T., Sassone-Corsi P. (2010) Cell Metab. 12, 509–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Duez H., Staels B. (2009) J. Appl. Physiol. 107, 1972–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lau P., Fitzsimmons R. L., Raichur S., Wang S. C., Lechtken A., Muscat G. E. (2008) J. Biol. Chem. 283, 18411–18421 [DOI] [PubMed] [Google Scholar]
- 32. Vieira E., Nilsson E. C., Nerstedt A., Ormestad M., Long Y. C., Garcia-Roves P. M., Zierath J. R., Mahlapuu M. (2008) Am. J. Physiol. Endocrinol. Metab. 295, E1032–E1037 [DOI] [PubMed] [Google Scholar]
- 33. McManus E. J., Sakamoto K., Armit L. J., Ronaldson L., Shpiro N., Marquez R., Alessi D. R. (2005) EMBO J. 24, 1571–1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Green C. B., Douris N., Kojima S., Strayer C. A., Fogerty J., Lourim D., Keller S. R., Besharse J. C. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 9888–9893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liu C., Li S., Liu T., Borjigin J., Lin J. D. (2007) Nature 447, 477–481 [DOI] [PubMed] [Google Scholar]
- 36. Sonoda J., Mehl I. R., Chong L. W., Nofsinger R. R., Evans R. M. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 5223–5228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hussain M. M., Pan X. (2009) Trends Endocrinol. Metab. 20, 177–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Scheving L. A. (2000) Gastroenterology 119, 536–549 [DOI] [PubMed] [Google Scholar]
- 39. Pan X., Hussain M. M. (2007) J. Biol. Chem. 282, 24707–24719 [DOI] [PubMed] [Google Scholar]
- 40. Pan X., Hussain M. M. (2009) J. Lipid Res. 50, 1800–1813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ouagued M., Saraux B., Girard-Globa A., Bourdel G. (1980) J. Nutr. 110, 2302–2309 [DOI] [PubMed] [Google Scholar]
- 42. Oishi K., Atsumi G., Sugiyama S., Kodomari I., Kasamatsu M., Machida K., Ishida N. (2006) FEBS Lett. 580, 127–130 [DOI] [PubMed] [Google Scholar]
- 43. Pan X., Zhang Y., Wang L., Hussain M. M. (2010) Cell Metab. 12, 174–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schlierf G., Dorow E. (1973) J. Clin. Invest. 52, 732–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hems D. A., Rath E. A., Verrinder T. R. (1975) Biochem. J. 150, 167–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Edwards P. A., Muroya H., Gould R. G. (1972) J. Lipid Res. 13, 396–401 [PubMed] [Google Scholar]
- 47. Escobar C., Díaz-Muñoz M., Encinas F., Aguilar-Roblero R. (1998) Am. J. Physiol. 274, R1309–R1316 [DOI] [PubMed] [Google Scholar]
- 48. Tsutsumi K., Inoue Y., Kondo Y. (2002) Biol. Pharm. Bull. 25, 1360–1363 [DOI] [PubMed] [Google Scholar]
- 49. Gimble J. M., Floyd Z. E. (2009) J. Appl. Physiol. 107, 1629–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Shimba S., Ishii N., Ohta Y., Ohno T., Watabe Y., Hayashi M., Wada T., Aoyagi T., Tezuka M. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 12071–12076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang N., Yang G., Jia Z., Zhang H., Aoyagi T., Soodvilai S., Symons J. D., Schnermann J. B., Gonzalez F. J., Litwin S. E., Yang T. (2008) Cell Metab. 8, 482–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bray M. S., Young M. E. (2009) Obes. Rev. 10, Suppl. 2, 6–13 [DOI] [PubMed] [Google Scholar]
- 53. Durgan D. J., Moore M. W., Ha N. P., Egbejimi O., Fields A., Mbawuike U., Egbejimi A., Shaw C. A., Bray M. S., Nannegari V., Hickson-Bick D. L., Heird W. C., Dyck J. R., Chandler M. P., Young M. E. (2007) Am. J. Physiol. Heart Circ. Physiol. 293, H2385–H2393 [DOI] [PubMed] [Google Scholar]
- 54. Tsai J. Y., Kienesberger P. C., Pulinilkunnil T., Sailors M. H., Durgan D. J., Villegas-Montoya C., Jahoor A., Gonzalez R., Garvey M. E., Boland B., Blasier Z., McElfresh T. A., Nannegari V., Chow C. W., Heird W. C., Chandler M. P., Dyck J. R., Bray M. S., Young M. E. (2010) J. Biol. Chem. 285, 2918–2929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sharma S., Adrogue J. V., Golfman L., Uray I., Lemm J., Youker K., Noon G. P., Frazier O. H., Taegtmeyer H. (2004) FASEB J. 18, 1692–1700 [DOI] [PubMed] [Google Scholar]
- 56. McGavock J. M., Lingvay I., Zib I., Tillery T., Salas N., Unger R., Levine B. D., Raskin P., Victor R. G., Szczepaniak L. S. (2007) Circulation 116, 1170–1175 [DOI] [PubMed] [Google Scholar]
- 57. Haemmerle G., Lass A., Zimmermann R., Gorkiewicz G., Meyer C., Rozman J., Heldmaier G., Maier R., Theussl C., Eder S., Kratky D., Wagner E. F., Klingenspor M., Hoefler G., Zechner R. (2006) Science 312, 734–737 [DOI] [PubMed] [Google Scholar]
- 58. Singer S. J., Nicolson G. L. (1972) Science 175, 720–731 [DOI] [PubMed] [Google Scholar]
- 59. Irvine R. F. (2002) Sci. STKE 2002, re13. [DOI] [PubMed] [Google Scholar]
- 60. Sinclair A. J., Attar-Bashi N. M., Li D. (2002) Lipids 37, 1113–1123 [DOI] [PubMed] [Google Scholar]
- 61. Hirsch E., Costa C., Ciraolo E. (2007) J. Endocrinol. 194, 243–256 [DOI] [PubMed] [Google Scholar]
- 62. Garbarino-Pico E., Valdez D. J., Contín M. A., Pasquaré S. J., Castagnet P. I., Giusto N. M., Caputto B. L., Guido M. E. (2005) Neurochem. Int. 47, 260–270 [DOI] [PubMed] [Google Scholar]
- 63. Garbarino-Pico E., Carpentieri A. R., Castagnet P. I., Pasquaré S. J., Giusto N. M., Caputto B. L., Guido M. E. (2004) J. Neurosci. Res. 76, 642–652 [DOI] [PubMed] [Google Scholar]
- 64. Marquez S., Crespo P., Carlini V., Garbarino-Pico E., Baler R., Caputto B. L., Guido M. E. (2004) FASEB J. 18, 519–521 [DOI] [PubMed] [Google Scholar]
- 65. Stavinoha M. A., Rayspellicy J. W., Hart-Sailors M. L., Mersmann H. J., Bray M. S., Young M. E. (2004) Am. J. Physiol. Endocrinol. Metab. 287, E878–E887 [DOI] [PubMed] [Google Scholar]
- 66. Goodwin G. W., Taylor C. S., Taegtmeyer H. (1998) J. Biol. Chem. 273, 29530–29539 [DOI] [PubMed] [Google Scholar]
- 67. Durgan D. J., Trexler N. A., Egbejimi O., McElfresh T. A., Suk H. Y., Petterson L. E., Shaw C. A., Hardin P. E., Bray M. S., Chandler M. P., Chow C. W., Young M. E. (2006) J. Biol. Chem. 281, 24254–24269 [DOI] [PubMed] [Google Scholar]
- 68. Bray M. S., Tsai J. Y., Villegas-Montoya C., Boland B. B., Blasier Z., Egbejimi O., Kueht M., Young M. E. (2010) Int. J. Obes. 34, 1589–1598 [DOI] [PMC free article] [PubMed] [Google Scholar]