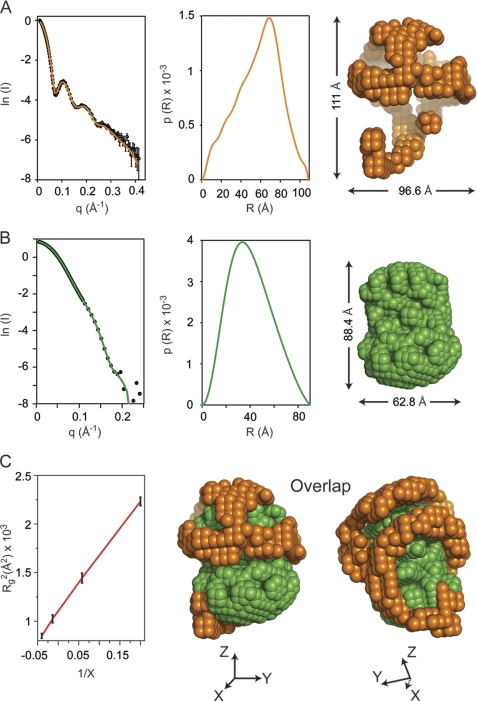
FIGURE 1.
Small angle neutron scattering results. A, neutron scattering results for sHDL in 12% D2O. Left panel, the scattering intensity as a function of the scattering vector q. The experimental data are plotted with open circles and have error bars attached. The experimental scattering curve displays oscillations with maxima at ∼0.1, 0.2, and 0.25 Å−1. The solid orange line is the scattering intensity calculated from the low resolution structure obtained by deconvoluting the experimental data. Middle panel, the distance distribution function, p(R), plotted as a function of the distance between scattering centers. The p(R) value was obtained by deconvoluting the experimental scattering intensities of sHDL in 12% D2O with the program GNOM. Right panel, low resolution structure of the protein component of sHDL obtained from the p(R) function (middle panel) with the program DAMMIN. B, neutron scattering results for sHDL in 42% D2O. Left panel, the scattering intensity (open circles) for sHDL in 42% D2O. The error bars are very small and only slightly visible at the center of the open circles. The solid green line is the scattering intensity produced by the low resolution structure of the lipid. Middle panel, the p(R) function for the lipid component of sHDL. Right panel, low resolution structure of the lipid component of sHDL obtained from the p(R) function (middle panel). C, left panel, the Stuhrmann plot gives the dependence of Rg2 (where Rg is radius of gyration) on the contrast level (X) in the SANS experiment. A linear graph (red line) indicates that the protein component is located on the outside, and the lipid component is inside as shown by the overlap of the low resolutions structures for the 12% D2O (orange) and 42% D2O (green) in the middle and right panels. X, Y, and Z represent the directions of the Cartesian coordinate system.