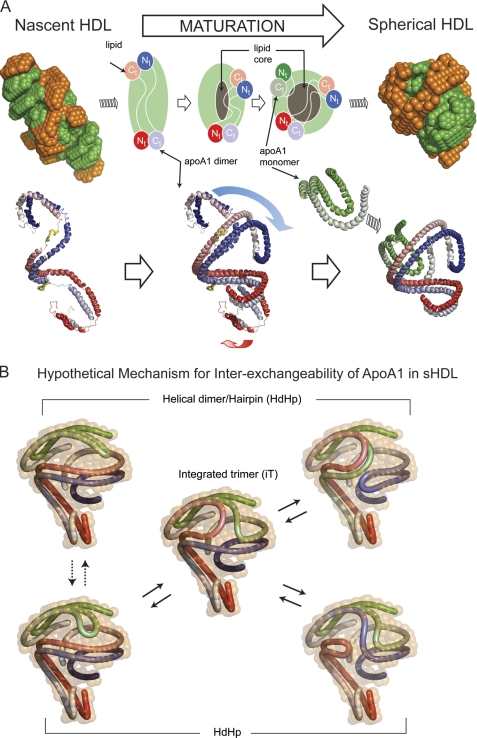
FIGURE 4.
Hypothetical mechanism of conformational change of apoA1 in nascent HDL during maturation into spherical HDL. A, left, superposition of the low resolution structures of protein (orange) and lipid (green) components of nascent HDL (top) and one particular orientation of the double super helix model of the protein component in nascent HDL (bottom left). The two apoA1 chains are gradient-colored red and blue, and the putative lecithin:cholesterol acyltransferase-binding loops (solar flares) are colored yellow. Middle, overlap of the middle domains of nascent HDL and sHDL apoA1 dimers. The overlapping suggests how the N and C termini of the apoA1 dimer in nascent HDL might swing and rearrange during particle maturation into what is found to be the dimer conformation in sHDL. The gray region at the middle of the particle represents the growing core of neutral lipids that accumulate during maturation. Right, superposition of the low resolution structures of protein (orange) and lipid (green) components of spherical HDL (top), and the resultant architecture of apoA1 trimer in spherical HDL (HdHp model, bottom right). B, hypothetical mechanism for apoA1 inter-exchange in sHDL. The hairpin and the apoA1 dimer are shown in different conformations that match the low resolution structure (12% D2O) of the protein. The different configurations suggest that it is reasonable to expect that because of particle dynamics the hairpin can bend such that it aligns its h5 domain with the h5 domain of the dimer (dotted double arrow lines) as a preamble for annealing with the dimer. Thus, the hairpin can exchange with one of the apoA1 monomers of the dimer through a transient integrated trimer-like configuration (center), generating other arrangements of the helical dimer/hairpin combination (solid double arrow lines). This protein reorganization mechanism makes all apoA1 monomers equivalent from an exchange point of view and can, in principle, lead to the integrated trimer (iT model) as illustrated on the right.