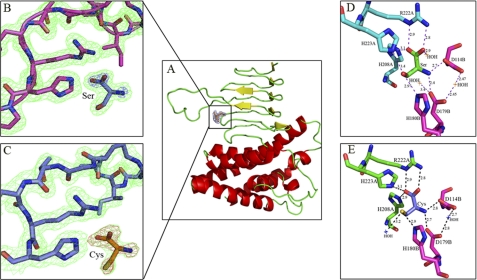
FIGURE 2.
A, EhSAT1 monomer shows bound ligand. The N-terminal region of the protein is arranged to form an α-helical-rich domain, and the C-terminal region is arranged to form a left-handed β-helical domain, characteristic of O-acetyltransferases. B, shown is a magnified view of the active site with bound Ser superposed with 2Fo− Fc electron density (green) at 1.2 sigma level in EhSAT1-Ser complex structure. (C) The magnified view of the active site with Cys superposed with 2Fo− Fc electron density (green color) at 1.4δ level in EhSAT1-Cys complex structure. In Fig. B and C, the bound amino acids were also superposed with 2Fo− Fc electron density map at 3δ level, shown in pink. D, interactions of l-Serine bound at the active site of the EhSAT1 molecule are shown. The complete active site is formed between two molecules of trimer; each molecule is shown in a different color. The carboxyl group of Ser makes a salt bridge with the side chain of Arg-222A and a hydrogen bond with side chain of His-208A and the water molecule. The hydroxyl group makes hydrogen bonds with the side chains of His-223A and His-180B. The amino group of serine makes a salt bridge with the carboxyl group of the Asp-114B and Asp-179B. A couple of water molecules also occupies the active site area, making hydrogen bonds with Asp-179B. E, interactions with the inhibitor l-cysteine bound at active site are shown. The position of the cysteine is identical to the serine in active site with minor changes. The carboxyl oxygen interacts with the amide group of the Arg-222A and also to His-223A and His-208A. The -SH group interacts with the His-180B, His-223A, and a water molecule. The -NH2 of cysteine bonds with the carboxyl groups of the Asp-114B and Asp-179B.