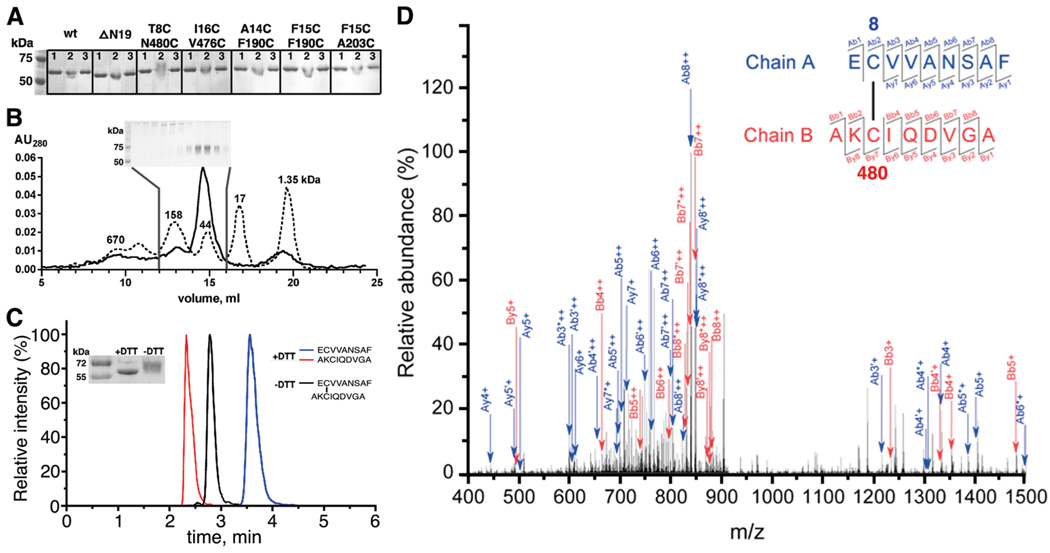
FIGURE 3.
GRK1535H6 T8C/N480C forms a disulfide bond between positions 8 and 480 under oxidizing conditions. (A) K3Fe(CN)6-treated GRK1535H6 T8C/N480C, but not wt, ΔN19, or other CC mutants, displays a delayed migration on nonreducing 10% SDS–PAGE that can be reversed by DTT: lane 1, before K3Fe(CN)6 treatment; lane 2, after K3Fe(CN)6 treatment; lane 3, after K3Fe(CN)6 treatment and then incubation with DTT. (B) GRK1535H6 T8C/N480C is monomeric after K3Fe(CN)6 oxidation. The elution profile from size-exclusion chromatography (Superdex 200, 25 mL) is shown as a solid line, and that of molecular mass standards is shown as a dotted line with numbers indicating molecular masses in kilodaltons. Nonreducing 10% SDS–PAGE analyzed fractions from the column are shown in the inset. Fractions of monomers show an upward gel shift, as seen in panel A. (C) Overlays of HPLC runs of peptidic fragments from oxidized GRK1535H6 T8C/N480C in the absence (black) or presence (red and blue) of DTT. The inset shows the degree of reduction via SDS–PAGE by application of DTT before tandem mass spectrometry analysis. (D) Identification of the Cys8–Cys480 disulfide bond by tandem mass spectrometry. The specific disulfide bond was verified on the basis of the MS2 spectra of the doubly charged ion at m/z 920.39. Ions used to identify fragments of chain A (EC8VVANSAF) are colored blue, whereas ions used to identify chain B (AKC480IQDVGA) are colored red. Product ions generated due to neutral losses are labeled using primes (loss of water) and asterisks (loss of ammonia). Further statistics for the confirmation of the disulfide-linked peptide by mass spectroscopy are provided in Table S1 of the Supporting Information.