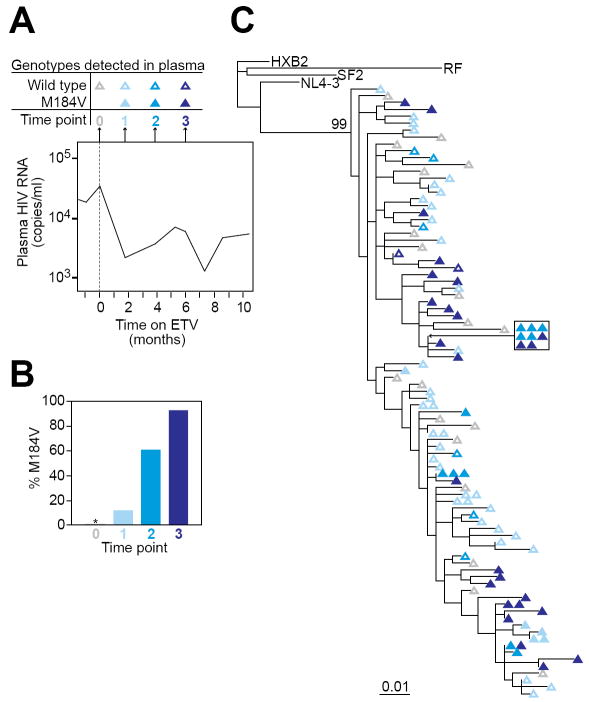
Figure 3.
Selection for the M184V mutation in HIV-1 RT in a patient on entecavir. (A) Time course of sampling for patient #1. Plasma was obtained for genotypic analysis on the day that entecavir was started (time point 0) and after 2, 4, and 6 months of entecavir monotherapy (time points 1, 2, and 3, respectively). The plasma HIV-1 RNA levels during this period are indicated in the graph. See Figure 1 for full details on the clinical course of patient #1. (B) Fraction of independent plasma virus isolates carrying the M184V mutation at time points 0, 1, 2, and 3. The number of independent clones analyzed was 19, 41, 18, and 27 for time points 0, 1, 2 and 3, respectively. * denotes no resistant mutations detected. (C) Maximum likelihood phylogenetic tree of wild type (open symbols) and M184V mutants (closed symbols) from time points 0, 1, 2, and 3. All isolates from patient #1 cluster together and are clearly distinct (bootstrap value =99) from reference clade B isolates obtained from other patients (HXB2, SF2, RF, and NL4-3)