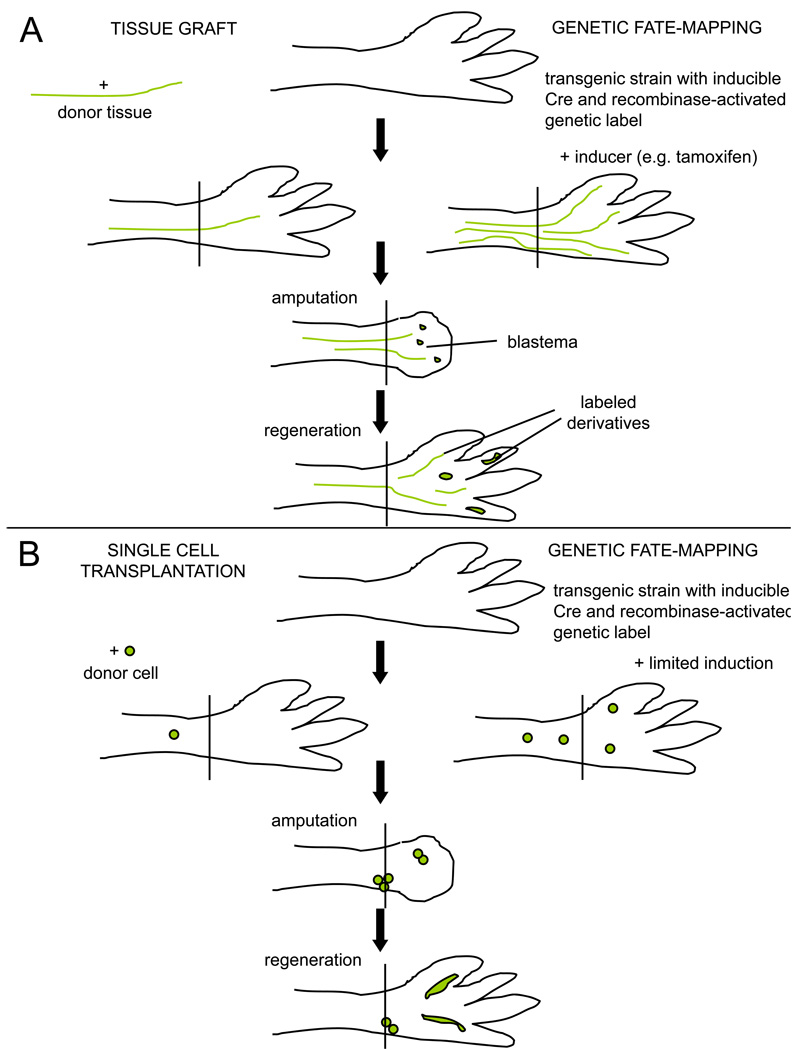
Box 2. Lineage tracing techniques used in regeneration studies.
A. Tests of tissue contributions. (Left) Tissue grafts are accepted well in salamanders, when performed either at the embryo or adult stage. A tissue type from a genetically distinct animal, e.g. a triploid animal or a transgenic animal expressing a fluorescent reporter gene in all cell types, can be dissected surgically and implanted into the intact limb of an unlabeled host. After amputation through the host limb, labeled tissues in the blastema and in the regenerate represent the derivatives of the graft. If multiple tissue contributions are detected after a transplant, issues of initial graft purity arise. (Right) Transgenic fate-mapping approaches typically use a cell type-specific promoter driving an inducible recombinase, e.g. a tamoxifen-inducible Cre fused to a mutated estrogen receptor, to trace the progeny of cells activating that promoter. After irreversibly labeling a limb cell type by tamoxifen injection, the limb is amputated and the blastema and regenerate assessed for labeled derivatives. A downside to this approach is that it relies on the availability and fidelity of a presumed tissue-specific promoter, relying on one marker for conclusions.
B. Clonal analyses can aid conclusions in lineage tracing experiments. (Left) A single labeled cell purified by flow cytometry is implanted into the intact host limb, or into the blastema (not shown). Labeled tissues in the regenerate can be traced back to that single cell, permitting rigorous tests of multipotentiality and self-renewal. (Right) A limited dose of tamoxifen will induce recombination in a small number of isolated cells, and presumably in those cells with relatively strong activation of the cell type-specific promoter driving the recombinase. Clones of cells in the regenerate can be assessed for cell type-specific markers to determine the tissue diversity of clonally related cells.