Fig. 1.
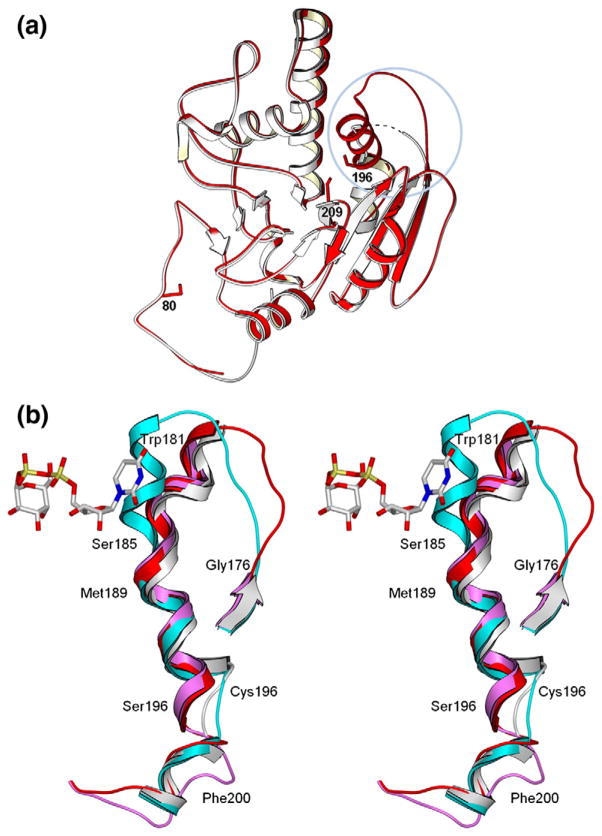
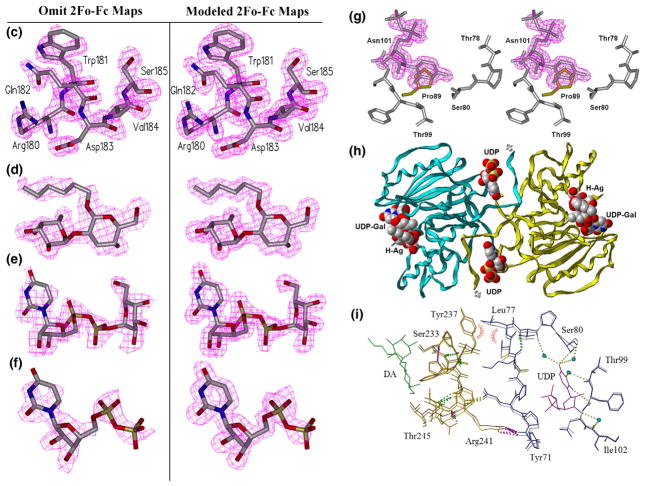
Effect of mutation on loop stability and on the second acceptor-dependent UDP binding site. (a) Secondary-structure overlap of wild-type GTB (white) and GTB/C80S/C196S/C209S (red) displaying the locations of the three Cys-to-Ser mutations and the internal disordered loop spanning residues 166–185 (circled). (b) Close-up of this loop displaying the open (white) and closed (cyan) conformations of wild-type GTB overlapped with GTB/C80S/C196S/C209S (red) and GTB/C80S/C196S (magenta), which display an ordered open conformation with UDP-Gal displayed in the classic ‘tucked under’ conformation described in Ref. 9. (Fo − Fc)-omit map generated after refinement of the initial molecular replacement solution and before modeling of ligand (left, contoured at 1.0 σ) and modeled 2Fo − Fc maps after the completion of refinement (right, contoured at 1.0 σ) of (c) the internal loop’s ordinarily disordered residues 180–185 from PDB 3I0H and from PDB 3I0L, (d) the deoxy-acceptor H antigen analog α-L-Fucp-(1 → 2)-β. -D-(3-deoxy)-Galp-OR (DA), (e) UDP-Gal observed in donor binding site, and (f) UDP observed at the dimer interface. (g) Stereoview of the UDP molecule observed at the dimer interface shown in its binding site with Pro89 of the symmetry-related molecule shown in yellow. (h) The dimer interface observed between [x,y,z] and [1 − x,y,1/2−z] z] 2-fold symmetry-related molecules showing the entire protein backbone of both dimer subunits and the observed bridging UDP molecules as well as UDP-Gal and DA in the active sites represented as van der Waals spheres. (i) Relative motion of the residues lying between the second UDP and acceptor binding sites of the unliganded and UDP + H mutant enzyme structures.