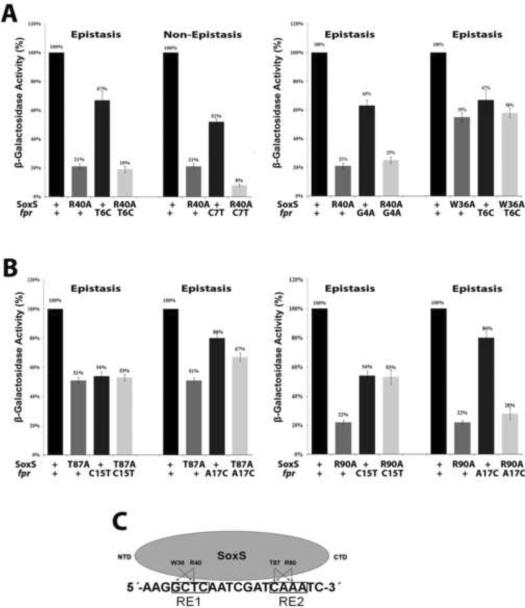
Figure 2.
Genetic epistasis experiments between single base pair substitutions of the soxbox of the fpr promoter and DNA binding mutants of SoxS. The sequence of the template strand of the fpr soxbox is shown in panel C; the invariant A of the soxbox is at the 5' end of the sequence. (A) (Left) Epistatic and non-epistatic interactions between the R40A substitution in the N-terminal HTH motif of SoxS and base pair substitutions T6C and C7T of RE 1, respectively. (Right) Epistatic interactions between the W36A and R40A substitutions in the N-terminal HTH motif of SoxS and substitutions T6C and G4A of RE 1, respectively. (B) (Left) Epistatic interactions between the T87A substitution in the C-terminal HTH motif of SoxS and base pair substitutions C15T and A17C of RE 2. (Right) Epistatic interactions between the R90A substitution in the C-terminal HTH motif of SoxS and substitutions C15T and A17C of RE 2. (C). A schematic representation of the interactions between amino acid residues of SoxS and nucleotide bases within the fpr soxbox, as determined by the genetic epistasis tests shown in (A) and (B). The epistasis data show that amino acids of the N- and C-terminal HTH motifs of SoxS contact one or two base pairs within RE 1 and RE 2, respectively, but whether the contacted base is on the template or non-template strand cannot be determined by these tests.