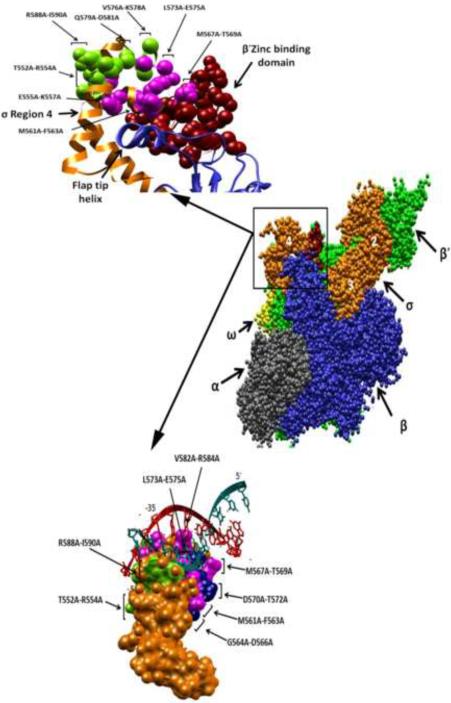
Figure 5.
Location of the equivalent positions of amino acids substituted with alanine in the trialanine scanning library of σ70 R4 on the crystal structures of holo-RNAP from Thermus thermophilus10 and on the crystal structure of Thermus aquaticus σA bound to DNA containing a consensus −35 element68. Only substitutions that reduce SoxS-dependent transcription activation of class I promoters or confer a defect in growth in liquid medium or on plates are identified. A space-filling model of the T. thermophilus RNAP holoenzyme is shown in the center, with the different subunits labeled and color-coded: σA subunit, orange; β subunit, blue; β' subunit, green; α subunit, grey; and ω subunit, yellow. The region displaying the interaction between σA R4 and the β flap-tip helix is enlarged in the upper left corner. The colors of the subunits are same as those in the model of holo-RNAP. Shown in magenta are the amino acid residues of σA that are equivalent to those within four (E555-K557, M561-F563, M567-T569 and L573-E575) of the five tri-alanine stretches of σ70 R4 that reduce SoxS-dependent transcription activation of class I promoters but have no effect on growth; the amino acids equivalent to those in the other stretch (V582-R584) do not appear in the crystal structure. The amino acids of σA that are equivalent to those within the four stretches of tri-alanine substitutions σ70 R4 that reduce SoxS-dependent transcription activation and also confer a defect in growth (T552-R554, V576-K578, Q579-D581 and R588-I590) are shown in green. Not appearing in this crystal structure are the amino acids of σA that are equivalent to those within the two tri-alanine stretches that confer a defect in growth (G564-D566, D570-T572) but not in SoxS-dependent transcription of class I promoters. The lower left corner displays the model of the crystal structure of σA R4 bound to DNA containing a −35 promoter hexamer. The amino acids residues of σA that are equivalent to those within four (M561-F563, M567-T569, L573-E575, V582-R584) of the five stretches of trialanine substitutions of σ70 R4 that reduce SoxS-dependent transcription activation of class I promoters but have no effect on growth are shown in magenta; the amino acids equivalent to those within the other stretch (E555-K557) do not appear in the crystal structure. The amino acids σA that are equivalent to two (T552-R554 and R588-I590) of the four stretches of trialanine substitutions that reduce transcription activation and also confer a growth defect are shown in green; the amino acids of σA that are equivalent to those within the other two tri-alanine stretches do not appear in this crystal structure. The amino acids of σA that are equivalent to those within the two tri-alanine stretches that confer a defect in growth (G564-D566, D570-T572) but not in SoxS-dependent transcription of class I promoters are shown in blue. The modeling was carried out with the Chimera molecular modeling software89, as described in Materials and Methods.