Fig. 1.
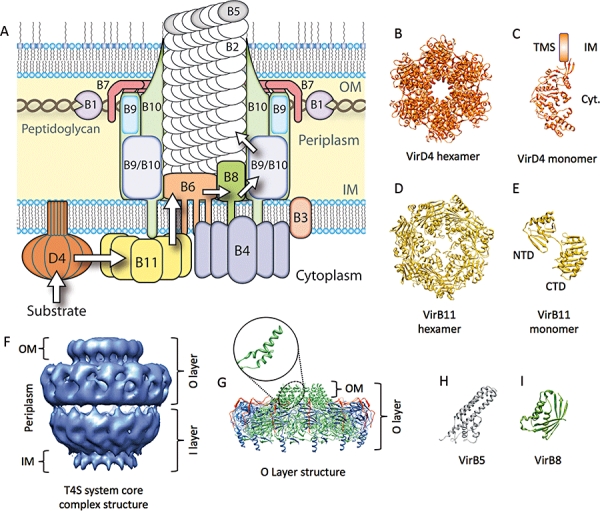
Schematic model of the VirB/D4 system of A. tumefaciens and structures of T4SS components determined to date.
A. The experimentally predicted locations of VirB/D4 components of A. tumefaciens. Arrows indicate the sequential translocation steps identified during substrate secretion through the VirB/D4 system in A. tumefaciens (Cascales and Christie, 2004).
B. Crystal structure of the TrwB hexamer, VirD4 homologue of the E. coli conjugative plasmid R388 (Gomis-Ruth et al., 2001).
C. Single subunit of the TrwB hexamer with modelled plausible location of the N-terminal transmembrane domain.
D. Crystal structure of the HP5025 hexamer, VirB11 homologue of H. pylori (Savvides et al., 2003).
E. Single subunit of the HP0525 hexamer. The N-terminal (NTD) and C-terminal (CTD) domains are indicated.
F. Cryo-electron microscopy structure of the T4SS core complex of the E. coli conjugative plasmid pKM101 (Fronzes et al., 2009b), comprising the full-length TraN, TraO and TraF, which correspond to the VirB7, VirB9 and VirB10 homologues of A. tumefaciens respectively.
G. Crystal structure of the outer membrane complex, comprising the O-layer (Chandran et al., 2009). The inset shows the characteristic two-helix bundle of TraF that traverses the outer membrane. TraN, TraO and TraF are coloured red, blue and green respectively.
H and I. Crystal structures of TraC, the VirB5 homologue encoded by pKM101 (Yeo et al., 2003), and the periplasmic C-terminal domain of VirB8 from B. suis respectively (Terradot et al., 2005).
Colour-coding of subunits in (A) is preserved for the individual subunits shown in (B)–(E) and (G)–(I). OM, outer membrane; IM, inner membrane; TMS, transmembrane segment; Cyt., cytosol.
