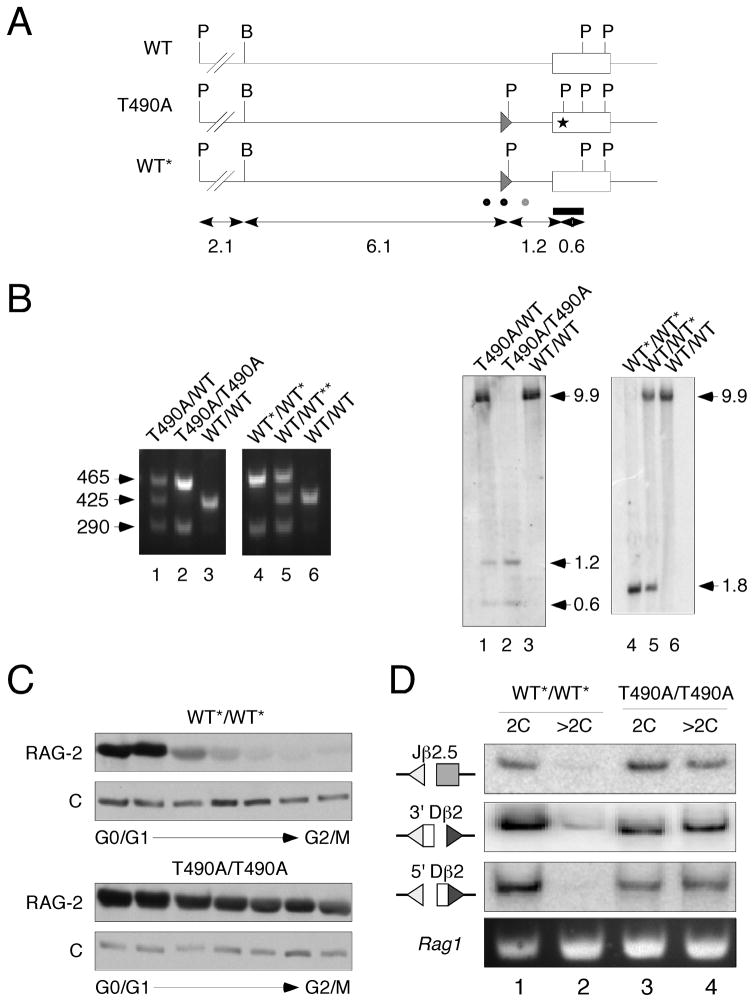
Figure 1.
Uncoupling of V(D)J recombination from cell cycle in Rag2(T490A) knockin mice. (A) Generation of Rag2(T490A) knockin mice. Top, wild-type Rag2 allele (wt); middle, neo-excised Rag2(T490A) allele (T490A); bottom, neo-excised, wild-type Rag2 allele (WT*). Open box, Rag2 coding exon. Direction of transcription is right to left. Filled triangle, loxP site; P, PstI; B, BamHI; asterisk, location of nucleotide substitutions specifying the T490A mutation and a novel PstI restriction site. Bar, probe used to assay recombinants by Southern hybridization. Dots below indicate positions of forward (black) and reverse (gray) PCR primers used for genotyping. Sizes of restriction fragments, in kb, are indicated below. (B) Assay for wild-type (WT), neo-excised wild-type (WT*) and neo-excised mutant (T490A) alleles. Genomic DNA was screened by PCR (left panels) with primers indicated in (A). Sizes of amplicons are indicated in bp. Genotypes were confirmed by Southern hybridization (right panels). Genomic DNA was digested with PstI and hybridized to the probe indicated in (A). Sizes of products are indicated in kb. (C) Cell cycle independence of RAG-2 accumulation in RAG-2 T490A mice. Thymocytes from wt*/wt* or T490/T490A mice were fractionated by centrifugal elutriation. Progression from 2C (G0-G1) to 4C (G2-M) DNA content is indicated below. Fractions were assayed for RAG-2 by immunoblotting. C, invariant, cross reactive species, used as an internal control. (D) Accumulation of RSS breaks is uncoupled from cell cycle in RAG-2(T490A) mice. Thymocytes from neo-excised wild-type (WT*/WT*) or mutant (T490A/T490A) mice were sorted into fractions of 2C or >2C DNA content. Double-strand DNA breaks at Jβ2.5, 3′Dβ2 and 5′Dβ2 RSSs (indicated at left) were assayed by LM-PCR amplification of genomic DNA. A portion of the Rag1 gene was amplified in parallel to monitor the amounts of genomic DNA in each reaction.