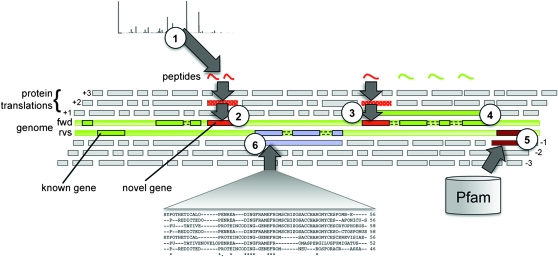
Figure 6.—
A cartoon representing the gene detection pipeline. The entire genome is translated in all three forward and reverse reading frames and segmented at putative stop codons. Data from different sources are compared to these translations and mapped back to the genome. Matching features that do not overlap with existing genes are used to infer novel genes and/or to refine gene structures. (1) MS/MS data searched against all putative protein sequences that could be expressed from the genome identify novel protein-coding genes (2) and refine 5′ and 3′ ends of others (3). MS/MS data matching pseudogenes, and genes annotated as noncoding, confirm translation and lead to altered annotation (4). Pfam searches against six-frame translations identify additional protein domains (5). Comparative genomic searches between whole-genome protein translations of six fungal species to S. pombe are used to infer novel protein coding genes (6).