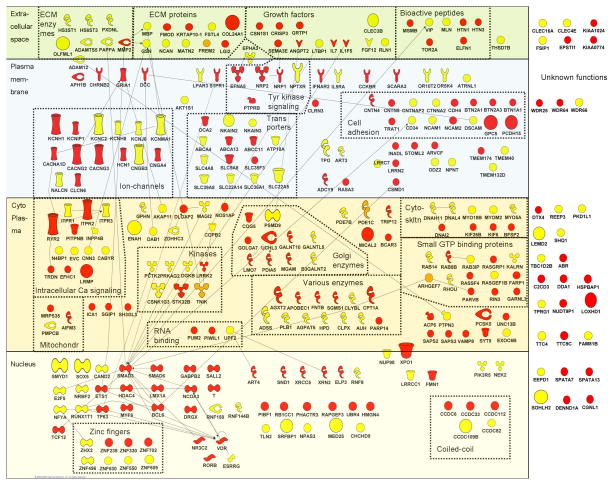
Figure 1. Connectivity diagram of the proteins encoded by the top genes (SNP with p-value <.001 in or close to gene) from the meta-analyses for AD and ND.
SNPs with a p-value < 0.001 in the meta-analyses of AD (N=515 SNPs) and ND (N=386) were selected. Of these, 261 AD and 213 ND SNPs were located in or within 10,000 bp of 200 and 165 genes, respectively. Those gene names were entered in the IPA database (Ingenuity Systems, release IPA 6.0) and 169 AD and 143 ND genes/gene-products were found in the database (gene names that were not mapped included loci named LOCxxx). We applied a network-based approach that grouped these genes by the biological functions, cellular locations, and possible interactions of their encoded proteins.
The proteins are represented as nodes and are displayed in various shapes that represent the functional class of the gene product (see symbol legend). Nodes were manually reorganized with the ‘pathway designer’ and the location of the nodes is based on the subcellular location of the gene products: extracellular, plasma membrane, cytoplasm, nucleus or unknown localization. Small symbols indicate genes with lowest SNP 0.001 < p-value < 0.0001, large symbols indicate genes with p-value < 0.0001. Depicted in red are the genes for AD, in yellow are those for ND and in orange those that are associated with both AD and ND. The resulting connectivity diagram showed several noteworthy groups of genes belonging to the same functional class (see dotted boxes), of which some are described in more detail in the text.