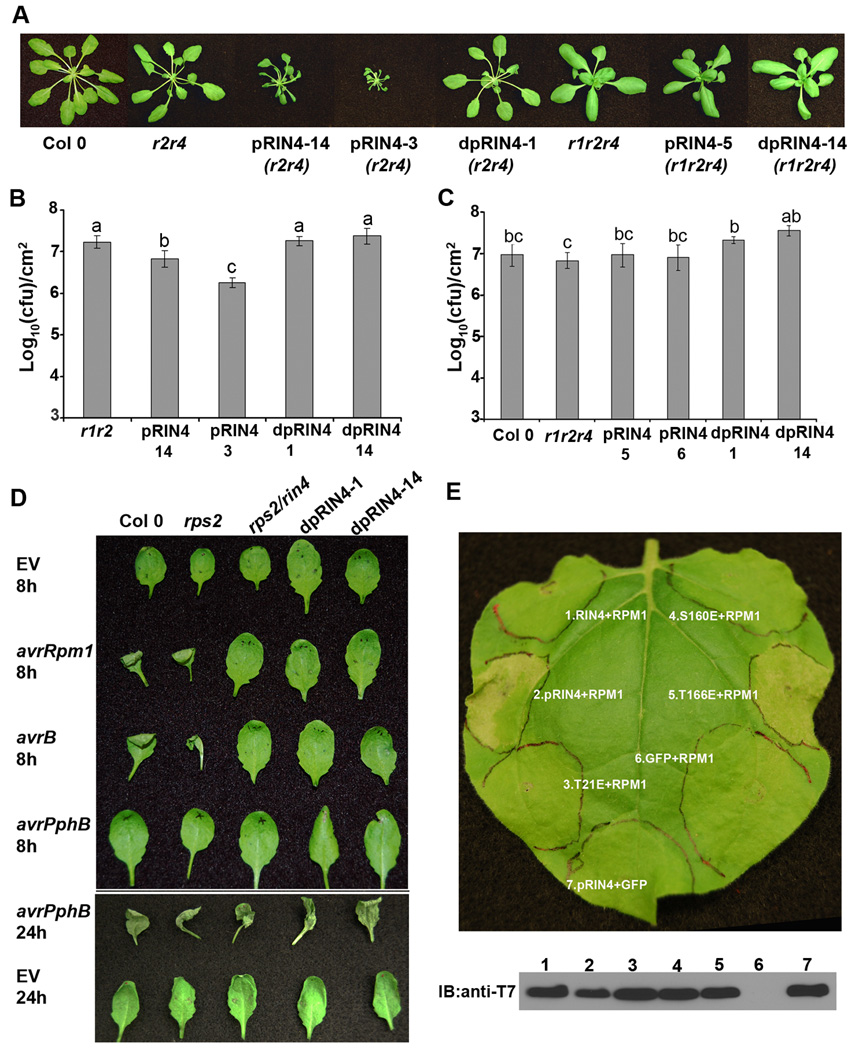
Figure 4. RPM1 recognizes phosphorylated RIN4.
(A) RIN4 phosphorylation mimics induce RPM1-dependent dwarfism. npro∷pRIN4 (T21E/S160E/T166E) and npro∷dpRIN4 (T21A/S160A/T166A) were transformed into rpm1/rps2 (r1/r2), rps2/rin4 (r2/r4) and rpm1/rps2/rin4 (r1/r2/r4). Representative pictures were taken from 4 week old T1 plants. (B) and (C) Four-week-old Arabidopsis plants were spray inoculated with 1×109 cfu/ml of Pst DC3000. Growth curve analysis was performed 4 days post inoculation. Statistical differences were detected by Fisher’s LSD, alpha = 0.05. Experiments were performed on homozygous T3 lines in the rps2/rin4 background and T2 lines in the rpm1/rps2/rin4 background. Transgenic lines originate from the T1 plants as indicated in (A). Error bars represent means ± standard deviation, n = 6. (D) Homozygous T3 transgenic lines expressing npro∷dpRIN4 in an rps2/rin4 background are no longer able to elicit HR in response to avrB or avrRpm1. Plants were syringe infiltrated with 2.5×107 cfu/ml of Pst DC3000 carrying empty vector, avrRpm1, avrB or avrPphB. Plants were photographed at 8h and 24h post-inoculation. (E) RPM1 recognizes RIN4 phosphorylation at T166. RPM1, T7-RIN4, T7-pRIN4(T21E/S160E/T166E), T7-RIN4(T21E), T7-RIN4(S160E), and T7-RIN4(T166E) were co-expressed in N. benthamiana using Agrobacterium-mediated transient expression. Leaf disks were sampled 40h post-infiltration for anti-T7 immunoblotting. Leaves were photographed for RPM1-induced HR 72h post-infiltration. The data shown are representative of three independent experiments with similar results.