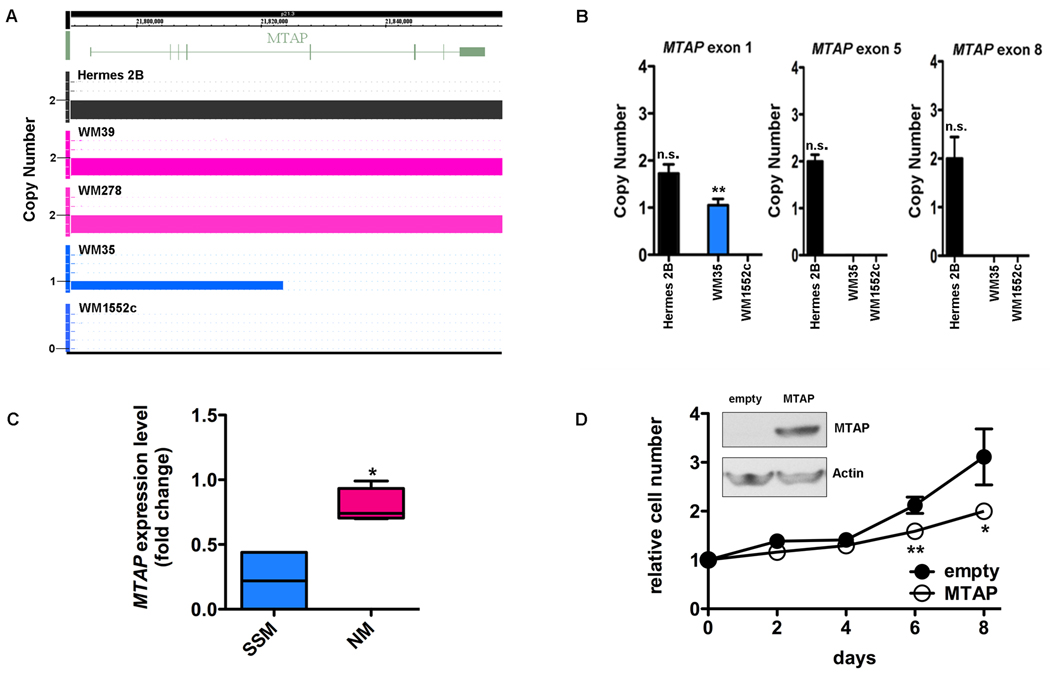
FIGURE 4.
A) SNP array in the region of MTAP shows that primary immortalized melanocyte Hermes 2B (control, black) and primary nodular melanoma cell lines (pink) have 2 copies of MTAP. There is a heterozygous deletion in SSM cell line WM35 (blue) spanning exons 1–4 and a homozygous deletion in exons 5–8. The deletion in WM1552c is homozygous and encompasses all exons. B) Genomic PCR of the MTAP gene in SSM cell line WM1552c confirms genomic losses in exons 1, 5, and 8, consistent with the large area of genomic deletion noted on SNP array. In SSM cell line WM35 (blue), only exons 5 and 8 are homozygously deleted which verifies the focal genomic loss detected using the SNP array. C) qRT-PCR verification of differential mRNA expression between NM (WM278, WM39, Lu451, SK-MEL-147) and SSM (WM1552c, WM35) cell lines in MTAP. D) Ectopic expression of MTAP in SSM cell line WM1552c by lentiviral infection (inset) results in decreased growth relative to control MTAP null cells. Mean ± SD, N=3. *P<0.05, **P<0.01, n.s.-not significant (relative to normal human genomic DNA in B).