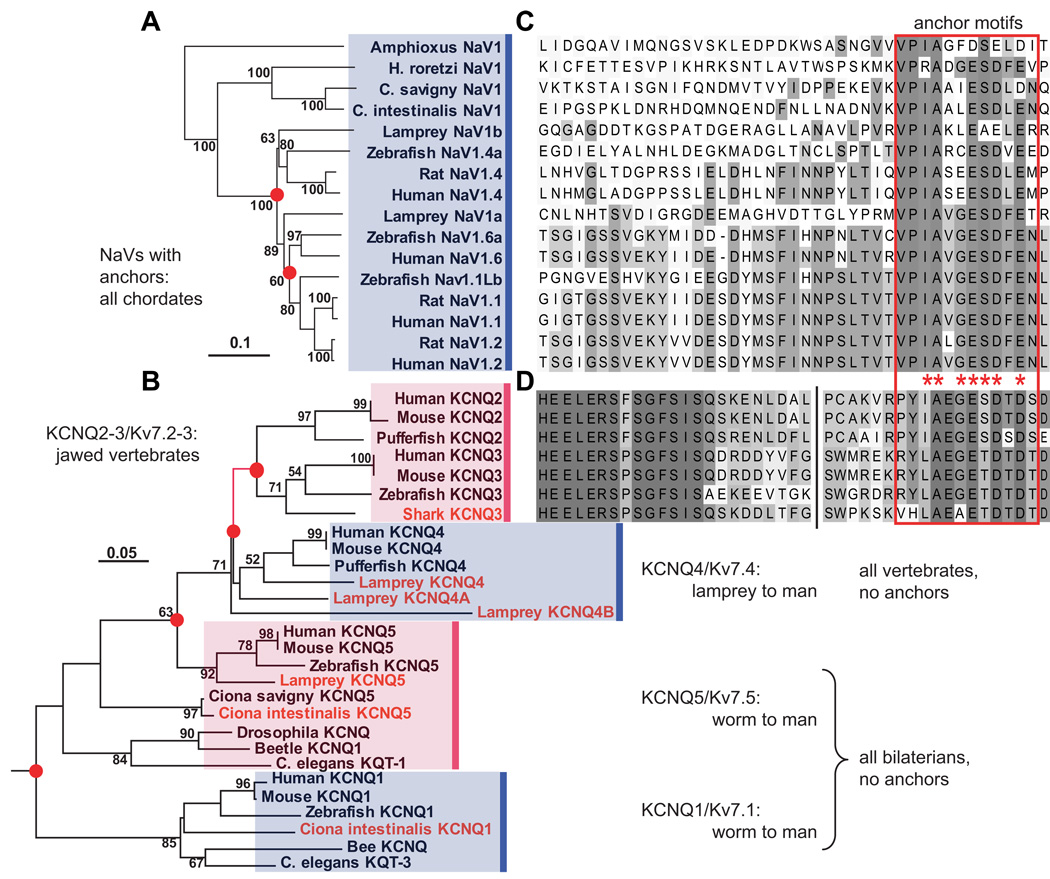
Figure 2. Phylogenetic analysis reveals that motifs for ankyrin-dependent axonal clustering evolved sequentially, first in chordates (NaV channels), then in jawed vertebrates (Kv7.2 and Kv7.3).
(A) Phylogram (minimal evolution) of NaV channels, showing that all vertebrate channels are derived from chordate NaV1. (B) Clustal alignment of NaV channel DII–DIII loop sequences, showing presence of anchor motifs in chordate NaV1 and all vertebrate channels. The anchor motifs are boxed (red). Shading indicates each residue’s conservation within a set of 28 chordate and non-chordate (not shown) aligned NaV sequences: bins represent ≤10, 11–20, 21–30, 31–45, 46–60, and 61–100 % conservation. (C) Phylogeny of KCNQ channels, based on analysis of derived amino acid sequences encoded on exons 5–7. Novel genes identified or cloned (see [12]) are shown in red. The branch marking the inferred first appearance of the KCNQ2/3 anchor motif is shown in red. (D) Alignment of KCNQ2 and KCNQ3 C-terminal intracellular sequences near the anchor motifs. Break (vertical black line) indicates location of 5–8 omitted, poorly conserved residues. The derived Kv7.2/Kv7.3 anchor motif sequences (red boxed region) are similar but non-identical to those of chordate NaV genes. Otherwise no similarity to the NaV DII–DIII loop sequence shown in B is present. Shading indicates conservation within the 7 KCNQ sequences aligned: shades represent ≤15, 15–30, 31–45, 46–60, 61–75, 76–90, and 91–100% conservation. In A and C, nodes are labeled with bootstrap values (>50%), nodes associated with gene duplications have red dots, and scale bars indicate substitutions per residue. After [12].