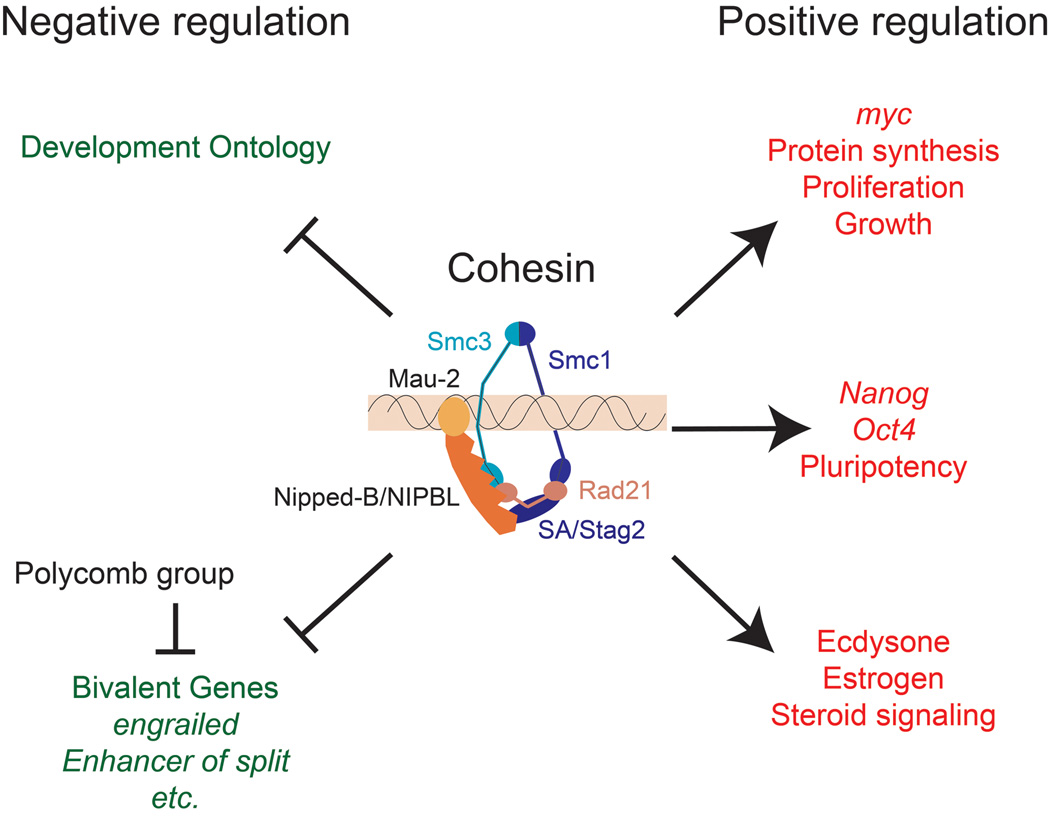
Figure 1.
Cohesin regulates expression of genes that control differentiation, morphogenesis, proliferation, and pluripotency. The cohesin complex consists of the Smc1, Smc3, Rad21 and Stromalin (SA)/Stag2 subunits [3]. Cohesin is thought to bind by encircling DNA, and binding requires the Nipped-B/NIPBL – Mau-2 complex, and ATPase activity in the Smc1 and Smc3 head domains that interact with Rad21. Both the genes that increase or decrease in expression with reduced Nipped-B/NIPBL or cohesin activity are enriched for cohesin-binding genes, indicating that cohesin both represses and facilitates transcription. Developmental processes are the top gene ontology categories for genes that increase in expression when cohesin or Nipped-B/NIPBL activity is reduced in Drosophila cells derived from central nervous system [15*], and mouse embryonic stem cells [28*]. The majority of the genes that show the largest increases in expression are bivalent, with both histone H3 lysine 4 and lysine 27 trimethylation, indicating that they are also partially repressed by Polycomb group silencing proteins. Cohesin directly promotes expression of genes in the ecdysone steroid hormone signaling pathway in salivary glands and the mushroom body γ neuron [13,14,24*] and the estrogen pathway in human breast cancer cells [27*]. Nipped-B/NIPBL and cohesin also directly facilitate expression of myc genes, which promote protein synthesis and cell proliferation in all species examined [12*,15*,26*,28*,30,31,34*], and pluripotency genes in mouse embryonic stem cells [28*].