Figure 1.
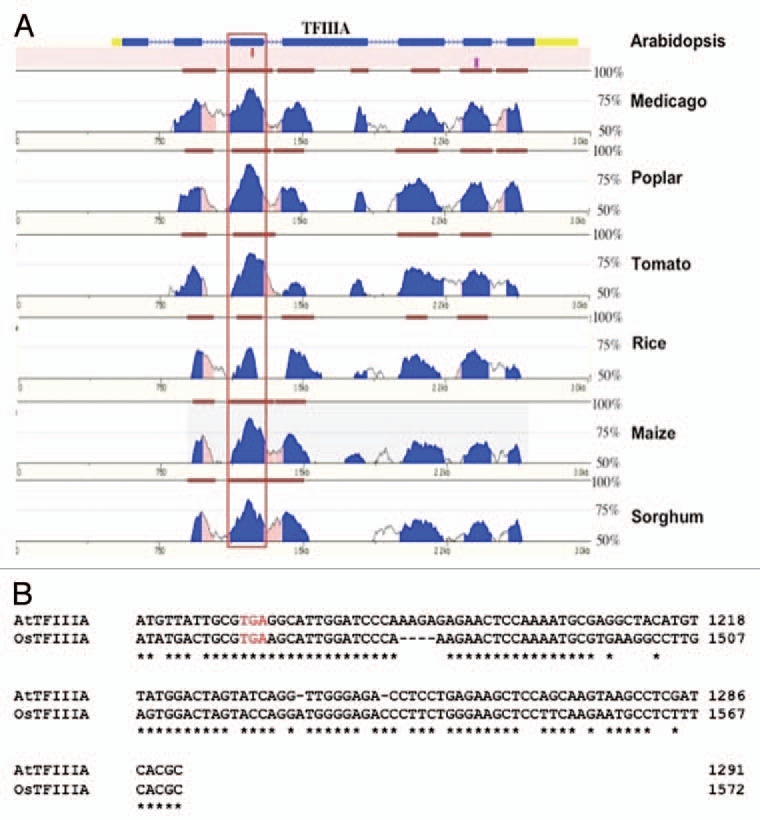
An example of sequence conservation between the alternative exons in a selection of plant TFIIIA genes. (A) Vista plots of pair-wise alignments between TFIIIA from Arabidopsis and its orthologue in medicago, poplar, tomato, rice maize and sorghum. The alternative exon (boxed) is the most conserved feature between Arabidopsis TFIIIA and its orthologues in other plant species. Levels of sequence identity between Arabidopsis TFIIIA and its orthologues in the six other plants species displayed are depicted as blue peaks. Brown bars signify segments that pass the alignment criteria of 70% identity over a window of 100 bp. (B) Detailed sequence alignment between the PTC containing alternative exon of Arabidopsis TFIIIA and one of the PTC containing alternative exons of rice TFIIIA—rice contains a duplication of the alternatively spliced exon (see text). Sequence identity across the alternatively splice exon is 74%. The in-frame stop codon introduced when this exon is included in the transcript is represented by red text.
