Figure 4.
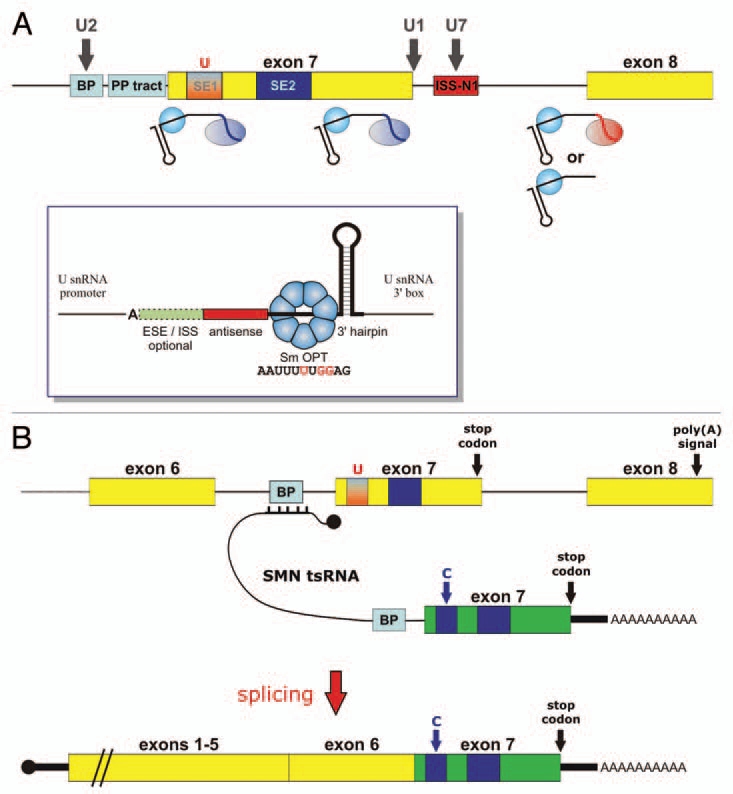
Scheme depicting splicing correction strategies for SMA based on in vivo expressed RNAs (A) SnRNA-based strategies. Bifunctional U7 Sm OPT derivatives have been designed to tether SR proteins to various positions in exon 7. Alternatively, U7 Sm OPT derivatives can mask the BP and 3′ SS of exon 8, either with or without tethering hnRNP A1. Shown with dark grey vertical arrows are snRNA-based strategies that were either not successful (U2 snRNA fully complementary to BP upstream of exon 7), inhibitory (U7 Sm OPT targeting ISS-N1) or toxic to cells (U1 snRNA fully complementary to the exon 7 5′ SS). Inset: Basic structure of a U7 Sm OPT derivative. The important elements are (from 5′ to 3′): the antisense sequence, the Sm OPT site capable of assembling with a heptameric Sm core of the standard Sm proteins (nucleotide changes respective to wild-type U7 snRNA are shown in red), and a 3′-terminal hairpin which stabilises the RNA. Splicing enhancer or silencer sequences can be added at the 5′ end to generate bifunctional U7 snRNAs. Note that transcription from a U snRNA promoter is important to allow efficient 3′ end formation at the U snRNA 3′ box and assembly into a snRNP particle. Moreover, mammalian U7 snRNAs start with an adenosine residue. (B) Trans-splicing strategy. A SMN-specific trans-splicing RNA (tsRNA) will bind to the BP/3′ SS region upstream of SMN2 exon 7 (for simplicity only the BP is shown) and will contain a strong BP/3′ SS leading into a SMN1-specific version of exon 7 (shown in green) and ending in a poly(A) tail downstream of the stop codon. After splicing, this SMN1-specific exon 7 will be fused to the body of the endogenous SMN2 mRNAs containing exons 1–6. Black knobs indicate the cap structures at the 5′ ends of the RNAs involved (see main text for refs.).
