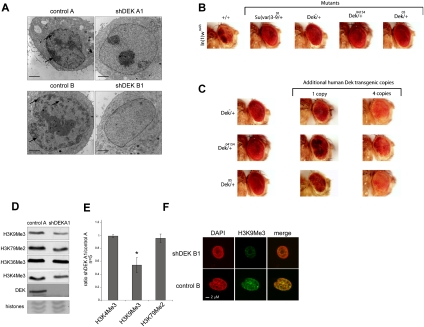
Figure 2.
DEK is a Su(var) (A) TEM reveals a global decrease in heterochromatic areas upon DEKkd. Cell lines were analyzed by standard TEM. Arrows highlight selected electron-dense heterochromatic areas. Bar, 2 μm. (B) Loss of a dose of Dek has a marked effect on white-mottled variegation in the eyes of male flies. Genomic deletion that includes the Dek coding region (Dek+/−) and P-element insertions into DEK (Dek+/04154 and Dek+/05) suppresses white-mottled variegation. The suppression was comparable with mutations in the well-characterized Su(var) KMT1 A/B [Su(var)3–9]. Representative fly heads were photographed 5–7 d after hatching. In(1)wm4h is the control fly. (C) The loss of a dose of Dek can be compensated by human DEK transgenes. The suppression of PEV in In(1)wm4h flies by Dek mutations carrying a heterozygous loss of Dek can be compensated by human DEK transgenes. (D) Assessment of epigenetic histone marks was performed by immunoblotting with modification-specific antibodies. (E) Average values from five independent experiments, as performed in D, are displayed and represent the ratio of signals obtained from total H3 amounts versus modification-specific signals. Error bars indicate ±SD. (*) P < 0.005. P-value was obtained by two-tailed Student's t-test, comparing H3K9me3 intensity in DEKkd cells with that in control cells. (F) IF staining with H3K9me3-specific antibodies was performed by confocal microscopy (see also Supplemental Fig. S5A).