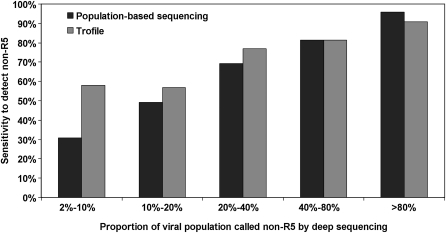
Figure 1.
Sensitivity of population-based V3 sequencing and original Trofile assay with 454 genotyping as the reference. The ability of the population-based V3 sequencing (black bars) and original Trofile (gray bars) to identify screening samples as having non–CCR5-using (non-R5) virus that 454 genotyping had identified as having ≥2% non-R5 virus, stratified by different proportions of non-R5 virus identified in the 454 genotyping result. Both alternative assays seemed to have decreased sensitivity for non-R5 human immunodeficiency virus (HIV) when such variants were present at lower proportions of the viral population. When non-R5 virus was present at 2%–10% according to 454 genotyping, 31% (69 of 224) and 58% (130 of 224) of the samples were also identified as having non-R5 virus by population-based sequencing and Trofile, respectively. These were 49% (57 of 116) and 57% (66 of 116) in the 10%–20% group; 69% (81 of 117) and 77% (90 of 117) in the 20%–40% group; 81% (118 of 145) and 81% (118 of 145) in the 40%–80% group; and 96% (180 of 188) and 91% (171 of 188) in the >80% group.