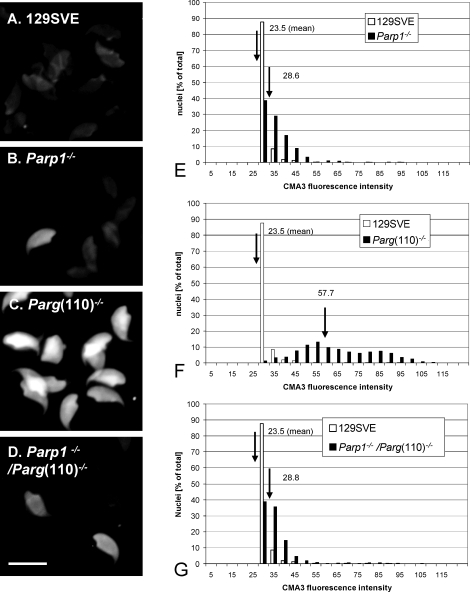
FIG. 2.
Low sperm chromatin quality in Parg(110)−/− mice. A–D) Chromomycin A3 (CMA3) intercalation into the DNA of epididymal spermatozoa was used as an indicator of incomplete chromatin maturation, as through insufficient DNA protamination or the presence of residual histone proteins in sperm. Bar = 10 μm. E–G) Quantitative measurement of CMA3 fluorescence revealed significantly (P < 0.001, ANOVA test) higher numbers of highly positive sperm nuclei in Parg(110)−/− mice (mean arbitrary fluorescence intensity F = 57.7, n = 12 animals) than in the wild-type 129 SVE mice (mean arbitrary intensity E–G = 23.5, n = 8 animals), Parp1−/− mice (mean arbitrary intensity E = 28.6, n = 4), or Parp1−/−/Parg(110) −/− animals (mean arbitrary intensity G = 28.6, n = 3 mice). Histogram analysis of data from a representative experiment (left panel, A–D) is shown in which labeled nuclei were categorized according to their measured fluorescence intensity and calculated as the percentage of that category relative to the total number of analyzed nuclei (n = 698 ± 77 nuclei/genotype). Statistical analyses showed that sperm from Parg(110)−/− mice were highly significantly different from all the other populations (P < 0.0001) whereas Parp1−/− and Parp1−/−/Parg(110)−/− populations were different from the wild type (P < 0.001, ANOVA test) but not from each other (P = 0.67).