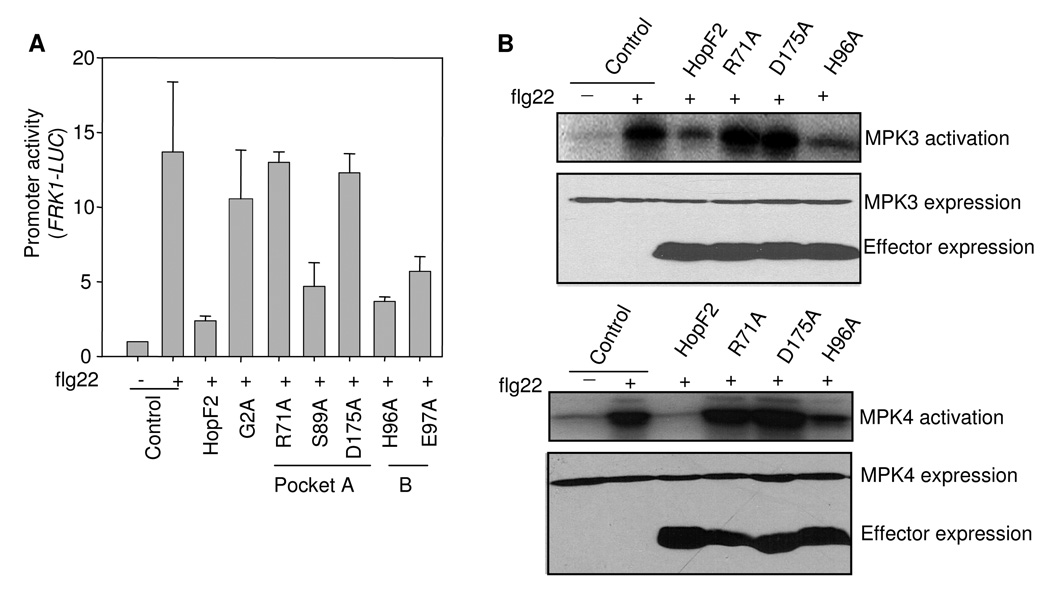
Fig. 4. Myristoylation and conserved surface residues of HopF2 are required for its MAMP-suppression function.
A. G2, R71 and D175 are required for HopF2 suppression of FRK1 induction. Protoplasts were co-transfected with HopF2 or its mutants and FRK1-LUC reporter, and incubated for 3 hr before treated with 10 nM flg22 for another 3 hr.
B. R71 and D175 are required for HopF2 suppression of MAPK activation. HA-tagged MAPK was co-expressed with HA-tagged HopF2 or its mutants for 6 hr before 1 µM flg22 treatment for 10 min. Kinase activity was detected by an in-vitro kinase assay (top). Protein expression is shown for MAPKs and effectors (bottom).