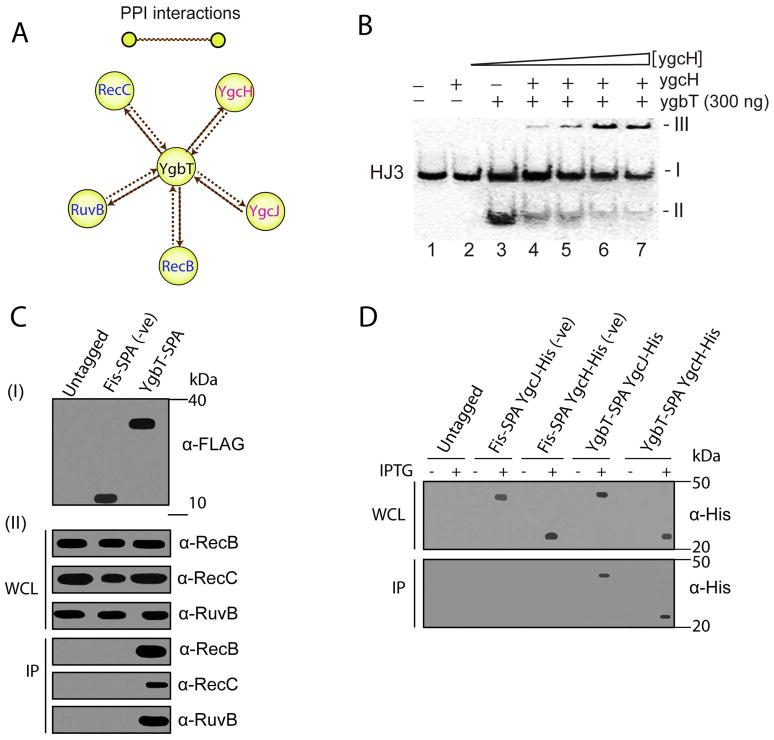
Fig. 4.
Physical interactions of YgbT with other E. coli proteins. (A) Interactions of YgbT with DNA repair/recombination and Cascade proteins validated by reciprocal SPA-tagging and purification. Yellow nodes represent tagged proteins used as “baits”, and brown edges represent protein-protein associations. (B) Purified YgcH (Cse3) inhibits the HJ cleavage activity of YgbT (native gel/autoradiography). The 5′-[32P]-labeled HJ3 substrate was pre-incubated for 2 min at room temperature with various amounts of YgcH (0, 0.1, 0.2, 0.3 and 0.4 μg) before the incubation with YgbT (0.3 μg, 45 min, 37 oC). The band-I represents the HJ3 substrate, the band-II - the HJ3 cleavage product (by YgbT), and the band-III (on the top) – the protein/DNA aggregates which are unable enter the gel. (C, D) Co-immunoprecipitation of YgbT with RecB, RecC, RuvB, YgcH, and YgcJ. Panel C show immunoblot analysis of the whole cell lysates (WCL) and anti-FLAG immunoprecipitates (IP) from the untagged DY330 strain (Panel C) or from strains expressing the SPA-tagged Fis or YgbT probed using RecB, RecC and RuvB antisera. Panels D show immunoblot analysis of the WCL and IP from the E. coli C41 (DE3) strain expressing a SPA-tagged Fis or YgbT with His6-tag YgcJ or YgcH probed using anti-His antibody.