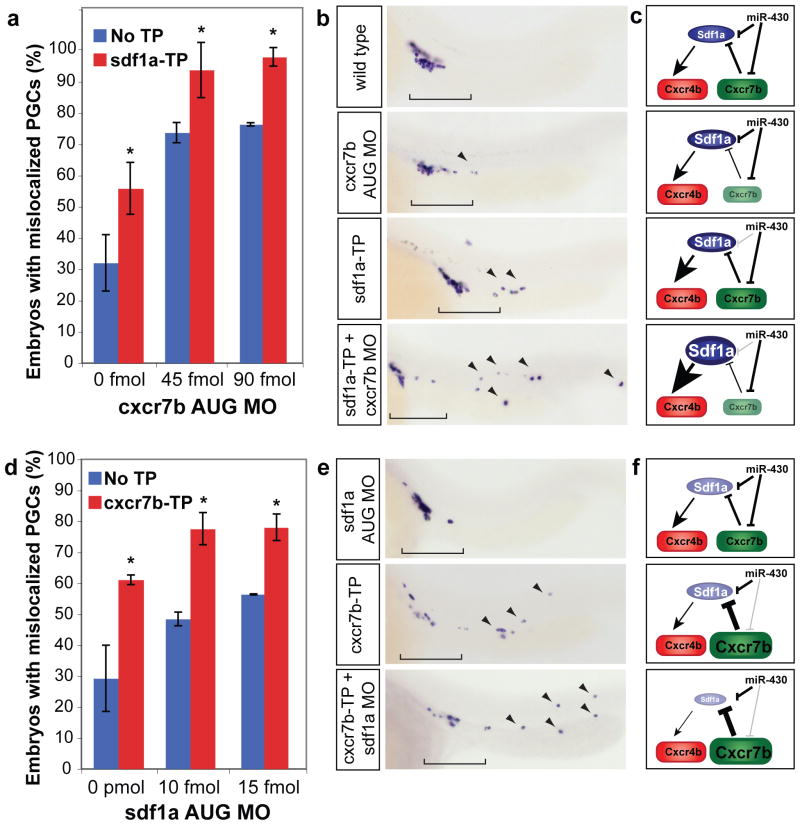
Figure 4.
miR-430 and Cxcr7b act in a functionally redundant manner to refine Sdf1a expression. (a, d) Quantification of PGC mislocalization. Embryos were injected at the one-cell stage with a morpholino targeting the start site (AUG MO) of cxcr7b (a) or sdf1a (d). These AUG MOs were injected at low concentrations, which were insufficient to completely knockdown the transcript and caused a weak mislocalization phenotype. (a) Co-injecting sdf1a-TP and cxcr7b AUG MO causes significantly more mismigration that injection of sdf1a-TP or the same amount of the AUG MO alone (*, p=4.08×10−3, sdf1a-TP + 45 fmol to 45 fmol alone; p=4.87×10−3, sdf1a-TP + 90 fmol to 90 fmol alone; two-tailed Fisher’s exact test), suggesting that miR-430 regulation of sdf1a mRNA can partially compensate for a reduction of cxcr7b. Similarly, co-injecting cxcr7b-TP and sdf1a AUG MO significantly enhances the mislocalization phenotype (*, p=8.93×10−4, cxcr7b-TP + 10 fmol to 10 fmol alone; p=0.016, cxcr7b-TP + 10 fmol to 10 fmol alone; two-tailed Fisher’s exact test), suggesting that regulation of cxcr7b by miR-430 prevents excessive clearance of the sdf1a. Data are shown as mean ± S.D. (b, e) nanos in situ at 24 hpf to visualize the location of germ cells. Brackets indicate correctly localized PGCs, and arrowheads show mislocalized cells. (c, f) Scheme representing the predicted effect of the experimental conditions on Sdf1a and Cxcr7b shown in panels (b and e). The added effect of removing miR-430 targeting and modulation by Cxcr7b supports a functional redundancy of miR-430 and Cxcr7b.