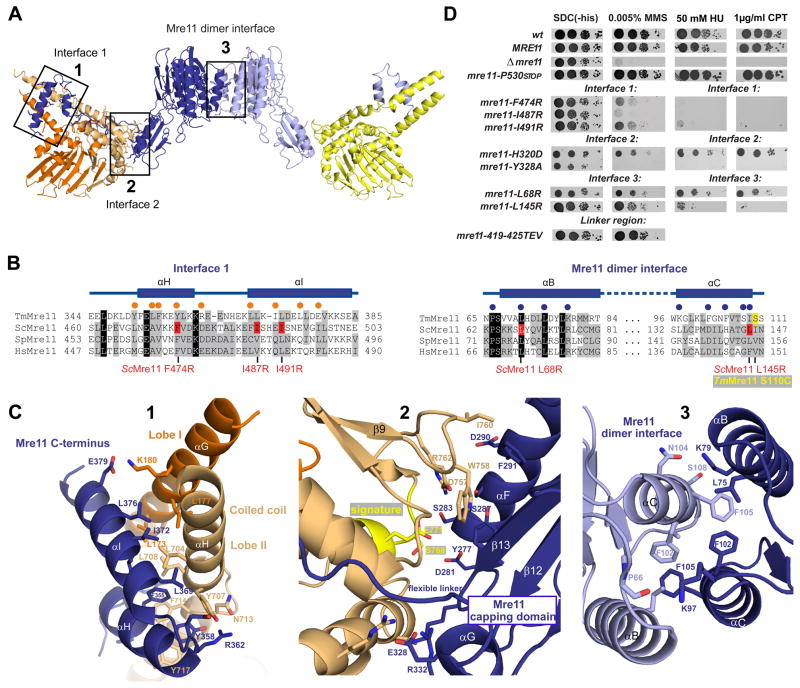
Figure 2. Details of Mre11:Rad50NBD and the Mre11 dimer interfaces.
A) Ribbon representation of the Mre11:Rad50NBD complex colored according to figure 1B. The boxed interfaces (1,2,3) are shown in C).
B) Sequence alignment of the Mre11 HLH domain (αH and αI) and the Mre11 dimer interface (αB and αC). Yeast mutations analyzed in D) are highlighted in red. Spheres represent residues implicated in Mre11HLH:Rad50NBD (orange) and Mre11:Mre11 interaction (blue).
C) Details of macromolecular interfaces. Selected side chains are shown as color-coded sticks and are annotated. 1) The Mre11 helix-loop-helix (HLH) motif (blue) and its interaction with the base of the Rad50 coiled coil (orange and yellow). 2) The interface between the capping domain of Mre11 (blue) and the opposite Rad50 (orange) region in close proximity to the signature motif (yellow). 3) The Mre11 dimerization interface.
D) Yeast survival assays by serial dilutions show that mutations in Mre11 predicted to affect interface 3 (L145R, L68R), mutations within the helix-loop-helix motif predicted to affect interface 1 (L474R, I487R, I491R), and mutations in the capping domain predicted to affect interface 2 (H320D, Y328A) result in sensitivity to Methyl-methanesulfonate (MMS), hydroxyurea (HU) and camptothecin (CPT). Changing the flexible linker residues (419–425 TEV) has no strong effect on MMS sensitivity. Left: SDC medium without histidine alone. Right: media supplemented with indicated concentrations of MMS, HU or CPT.
See also Figure S2.