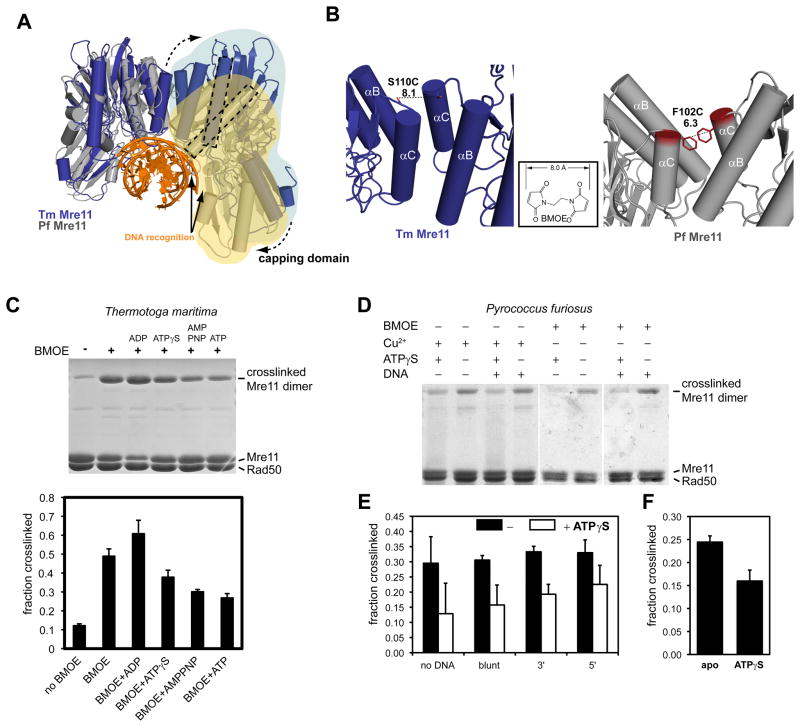
Figure 5. ATP binding to Rad50 changes the Mre11 dimer interface.
A) Comparison of the nuclease dimer of DNA bound PfMre11 (grey, yellow, orange) with that of TmMre11 (blue, lightblue). Both complexes are superimposed via the left protomer. In T. maritima, the right protomer has to undergo a substantial rigid body motion to adopt the same DNA binding contacts (solid arrows) and conformation as seen for PfMre11 (dashed arrow). This can be seen by the different orientations of the minor groove binding helix (dashed rectangles).
B) To probe for conformational changes, we mutated S110C in TmMre11 (left panel) and F102C in PfMre11 at a position that is expected to undergo movements when switching between different pivot angles. The distances are suitable for crosslinking with a short bifunctional sulfhydryl directed crosslinker (inset: BMOE), or by forming disulfide bonds.
C) Crosslinking analysis of TmMre11 dimer structure. Upper panel, non-reducing Coomassie stained SDS PAGE of TmMRNBD crosslinked under different conditions (lane labels see lower panel); Lower panel, quantification of the crosslinking efficiency (mean +/− standard deviation of three independent experiments) for different conditions and adenosine nucleotides. A crosslinked band corresponding to the Mre11 dimer in the absence of BMOE crosslinker indicates disulfide bond formation.
D) Coomassie stained non-reducing SDS PAGE of crosslinked PfMRNBD, showing different conditions and replicates. Cu2+ was used to increase disulfide bond formation.
E) and F) Quantification of crosslinking efficiency by BMOE (E) or disulfide bonds (F). Error bars depict standard deviations. In F) the effect of ATPγS is tested. In E) the effect of dsDNA 50mer with either blunt ends or 3′ or 5′ 10 dT ssDNA overhangs is shown. Black bars are without ATPγS, white bars with ATPγS.