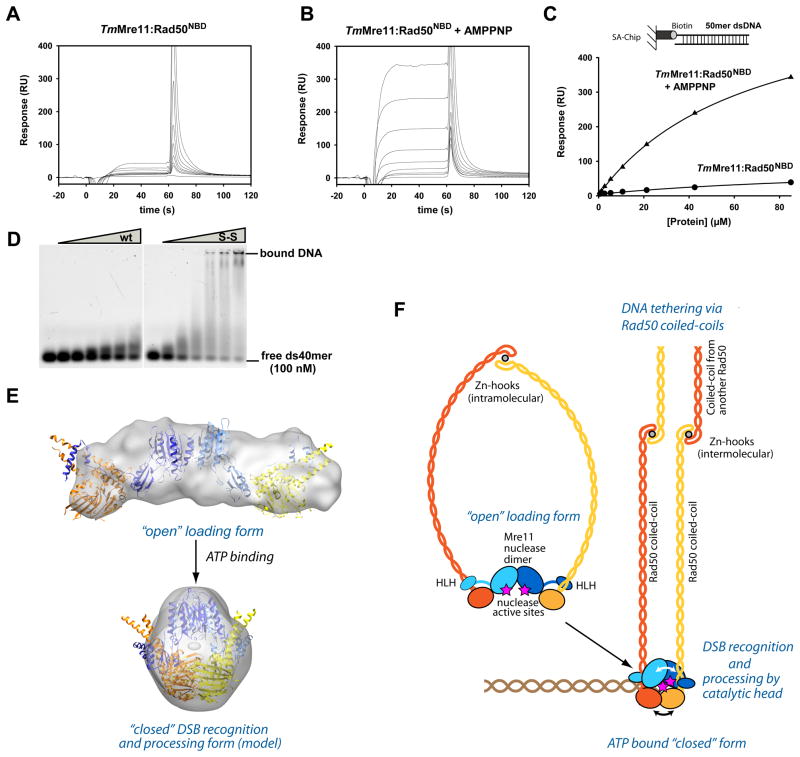
Figure 6. ATP stimulated dsDNA binding and clamp model for DSB sensing.
A) and B) Surface Plasmon sensograms for binding of TmMre11:Rad50NBD to 50mer dsDNA in absence (A) and presence of AMPPNP (B).
C) Corresponding binding curves reveals AMPPNP stimulated binding to dsDNA. AMPPNP also increases hairpin recognition (Fig. S6A) and we also observe increased binding affinity of the protein with crosslinked Rad50 NBDs (ATP mimic state) (data not shown).
D) Electrophoretic mobility shift assays show that the MRNBD,H830C,D804C complex with a disulfide bond between the NBDs (S-S) has strongly increased dsDNA oligonucleotide binding affinity compared to the wildtype MRNBD complex (wt). Following concentrations of the protein (0, 1.0, 2.5, 5.0, 7.5, 10.0 and 15.0 μM respectively) were used.
E) Model for the closed, clamp like complex with engaged NBDs by combining X-ray structures and SAXS analysis. “Open” (experimental) and “closed” (rigid body docked model) forms are displayed with corresponding SAXS envelopes.
F) Proposed model for ATP dependent DSB sensing and processing by MR by formation of a transient clamp at DNA end structures.
See also Figure S5.