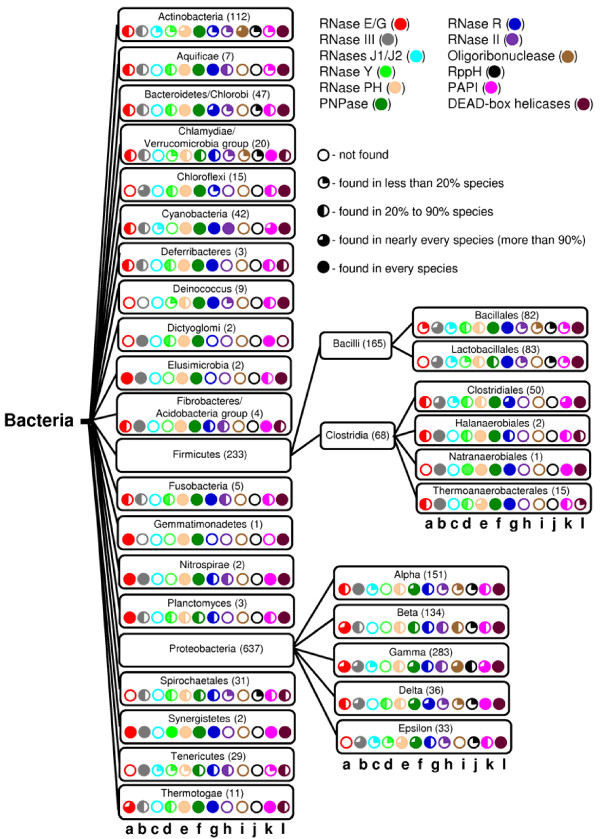
Figure 2.
The phylogenetic distribution of main ribonucleases (RNase E/G, RNase III, RNases J1/J2, RNase Y, RNase PH, PNPase, RNase R, RNase II, Oligoribonuclease) and ancillary RNA modifying enzymes (RppH, PAPI, DEAD-box helicases) involved in the disintegration and turnover of bacterial transcripts are indicated by colored filled circles (from 'a' to 'l', respectively). The percentage of organisms in each phylum/class of bacteria for which the presence of each particular enzyme has been predicted by searching the NCBI database is indicated by differentially colored circles. The data are compiled based on analysis of completely sequenced genomes (1217 complete genome sequences available by 4 November 2010). Draft assemblies of genomes and hypothetical proteins were excluded from the analysis.