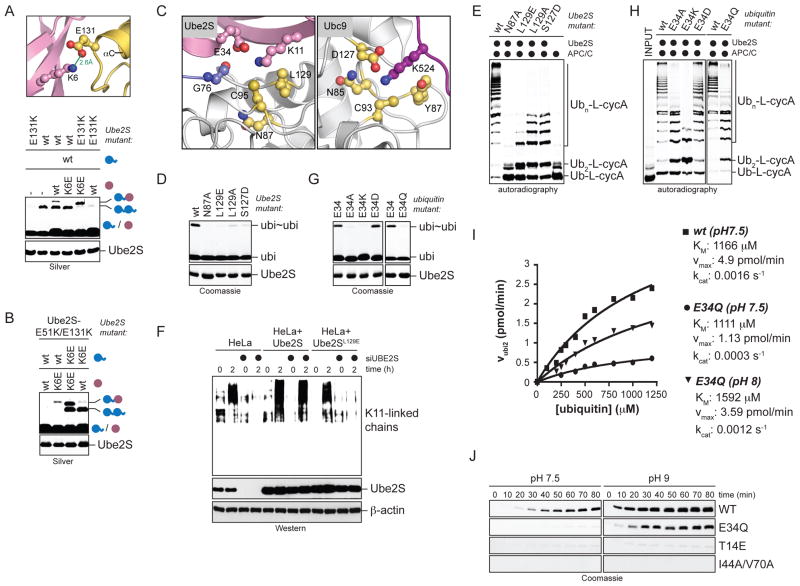
Figure 7. Substrate-assisted catalysis contributes to the K11-linkage specificity of Ube2S.
A. Charge swap analysis of the ionic contact between acceptor Lys6 and Glu131 of Ube2S. Ube2S or Ube2SE131K were mixed with ubiΔGG or ubiΔGG/K6E and reactions were analyzed by Silver staining. B. Ube2SE51K/E131K rescues mutation of Lys6 in both acceptor and donor ubiquitin. Lys6 was mutated in acceptor ubiΔGG (purple) or donor ubiquitin (blue), and ubiΔGG-ubi formation by Ube2S or Ube2SE51K/E131K was analyzed by Silver staining. C. Ube2S (left) and Ubc9 (PDB ID: 2GRN; right) show similar active site constellations. The highest scoring Ube2S-model of the HADDOCK run in the absence of ambiguous restraints is shown (Table S2 top; cluster 1, no 1). D. Candidate active site-residues are required for the activity of Ube2S to catalyze ubi2-formation (ubi~ubi), as analyzed by Coomassie staining. E. Active site-residues in Ube2S are required for chain elongation by APC/C. Ub-L-cycA was incubated with APC/CCdh1 and Ube2S mutants and analyzed by autoradiography. F. Leu129 is required for Ube2S-activity in vivo. HeLa cell lines expressing Ube2S or Ube2SL129 were tested for formation of K11-linked chains after endogenous Ube2S was depleted by siRNAs. K11-chain formation in cells arrested in prometaphase or exiting mitosis was monitored by αK11-Western. G. Glu34 of acceptor ubiquitin is required for K11-linkage formation. Ubiquitin mutants were incubated with Ube2S and analyzed by Coomassie staining. H. Glu34 of acceptor ubiquitin is required for chain elongation by APC/C and Ube2S. The modification of Ub-L-cycA by APC/C, Ube2S, and ubiquitin mutants was analyzed by autoradiography. I. ubiE34Q displays catalytic, but not binding defects. The rates of ubi2-formation at different concentrations of ubiquitin and ubiE34Q at the indicated pH were determined from two or three independent time-courses. Apparent kinetic constants were obtained by fitting the rate constants to a Michaelis-Menten equation. J. Rescue of ubiE34Q, but not other TEK-box or hydrophobic patch mutants, by increasing the reaction pH. Ubiquitin or indicated mutants were incubated with Ube2S at pH 7.5 (left) or pH 9 (right) and analyzed by Coomassie staining.