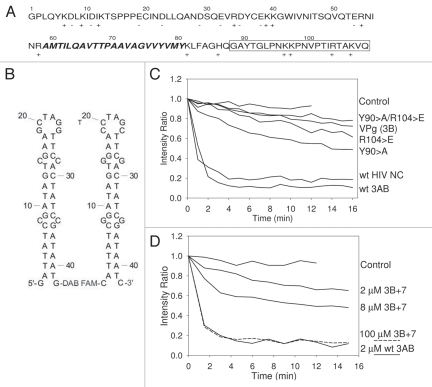
Figure 2.
(A–D) Depiction of 3AB sequence and results from FRET unwinding assay. (A) Amino acid sequence of poliovirus 3AB. Regions: 3A, 1–87: 3B (VPg), 88–109 (boxed); C-terminal cytoplasmic domain, 81–109 (referred to as 3B+7 in this proposal); membrane anchor domain, amino acids 59–80 (in bold italics). (B) Substrates for FRET unwinding assay. Complementary stem-loop forming oligos with 5′ fluorescing (FAM) or quenching DABCYL groups are shown. Hybridization leads to quenching of FAM fluorescence. Structures and ΔG values of −7.2 kcal/mol are as predicted from DNA fold.71 (C) FRET unwinding time course assay with wild type 3AB, 3AB-R104E, 3AB-Y90A, 3AB-R104E/Y90A, VPg (3B) and HIV NC, all at 2 µM. (D) FRET unwinding time course assay with wild type 3AB (2 µM) or 2, 8 and 100 µM 3B+7. Representative results are shown for all experiments and assays were repeated at least once and typically several times.