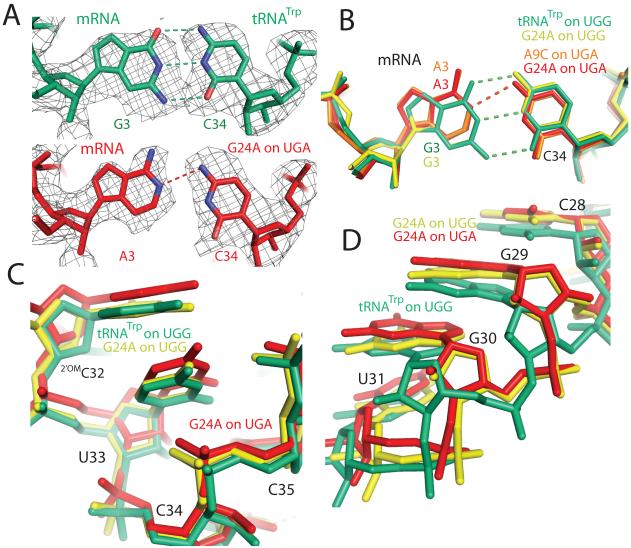
Figure 2.
Comparison of cognate and near-cognate structures in the decoding center. A) Unbiased Fo - Fc electron density (displayed at 1.3σ within 2 Å of EF-Tu) is shown for residues in the wobble position of the codon-anticodon helix. B) Binding of a near-cognate tRNA (A9C on UGA: orange, G24A on UGA: red), compared to a cognate tRNA (G24A on UGG: yellow, tRNATrp on UGG: green), results in a shift in both the tRNA anticodon and mRNA codon. C) The distortion in the anticodon caused by this mismatch is propagated three residues, resulting in a shift in the tRNA backbone for the G24A on UGA when compared to the G24A or tRNATrp on UGG. D) However this distortion does not continue past residue 31; by residue 28 the backbone of the G24A on UGA has converged with that of the G24A on UGG.