Figure 3.
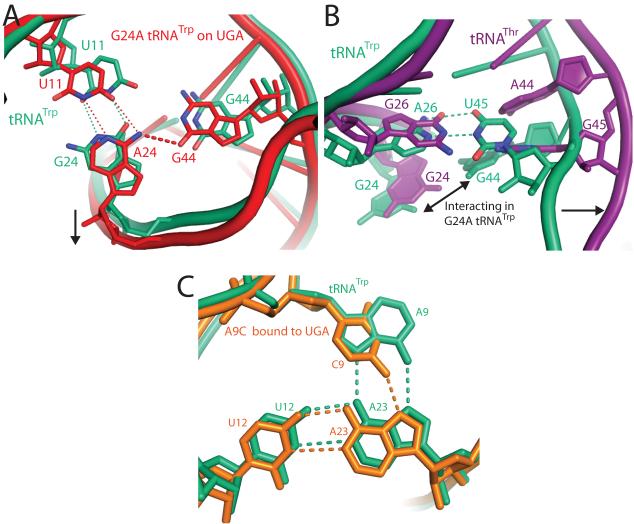
Miscoding by the G24A and A9C Trp-tRNATrp. A) Mutation of G24A (red) facilitates formation of a hydrogen bonding interaction that is not possible in native tRNATrp (green) between the exocylic amine of A24 and O6 of G44. This interaction is facilitated by a small shift in the RNA backbone between residues 20 to 30 and a commensurate movement between residues 9-16, B) The interaction between residues 24 and 44 in the G24A tRNATrp is only possible because of a base pair between residues A26-U45 in tRNATrp that dictates the backbone conformation in this region. In contrast, tRNAThr14(purple) contains two purines, G26 and G45, at these positions, which push the tRNA backbone apart and separate residues G24 and A44. That the A26-U45 base pair is not conserved even throughout bacterial tRNATrp further illustrates that evolution has fine-tuned each tRNA to find a unique solution to tRNA bending during decoding. (C) In native tRNATrp (shown in green) a base triple forms between residues 9:12:23, which is at the junction of three separate strands of the tRNA. Mutation of residue 9 to a cytosine (shown in orange) weakens both the packing and hydrogen-bonding of this base-triple, which could result in higher flexibility of the tRNA body and explain its ability to miscode.