Table 2. Classifying Angus samples.
| Angus | Panel 1 | Panel 2 | Panel 3 | |||
| Decision Tree Nodes |
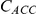
|
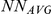
|
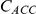
|
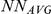
|
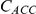
|
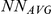
|
World  Bos taurus
Bos taurus
|
27/27 | 5.00 | 27/27 | 5.00 | 27/27 | 5.00 |
Bos taurus
 European taurine breeds European taurine breeds |
27/27 | 5.00 | 27/27 | 5.00 | 27/27 | 5.00 |
European taurine breeds  7 taurine breeds 7 taurine breeds |
27/27 | 5.00 | 27/27 | 5.00 | 27/27 | 5.00 |
7 taurine breeds  Angus-Red Angus Angus-Red Angus |
24/27 | 4.44 | 27/27 | 4.78 | 26/27 | 4.85 |
Angus-Red Angus  Angus Angus |
21/27 | 3.89 | 26/27 | 4.67 | 25/27 | 4.3 |
Predicting the breed of individuals in the Angus (ANG) bovine population using our PCAIM SNP panels P1, P2, and P3. A total of 27 ANG individuals were available in the Bovine HapMap dataset. The 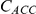 columns correspond to classification accuracy, expressed as the fraction of individuals that were assigned to the correct breed at the respective node of the decision tree, and
columns correspond to classification accuracy, expressed as the fraction of individuals that were assigned to the correct breed at the respective node of the decision tree, and 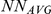 indicates the average number of correct neighbors at the same node of the decision tree. For example, at the seven-taurine-breeds node of the decision tree, 24 out of the 27 Angus samples were (correctly) predicted to be of Angus-Red Angus origin using Panel 1; at the same node, the ANG individuals had – on average – 4.44 neighbors from within the Angus breed.
indicates the average number of correct neighbors at the same node of the decision tree. For example, at the seven-taurine-breeds node of the decision tree, 24 out of the 27 Angus samples were (correctly) predicted to be of Angus-Red Angus origin using Panel 1; at the same node, the ANG individuals had – on average – 4.44 neighbors from within the Angus breed.
