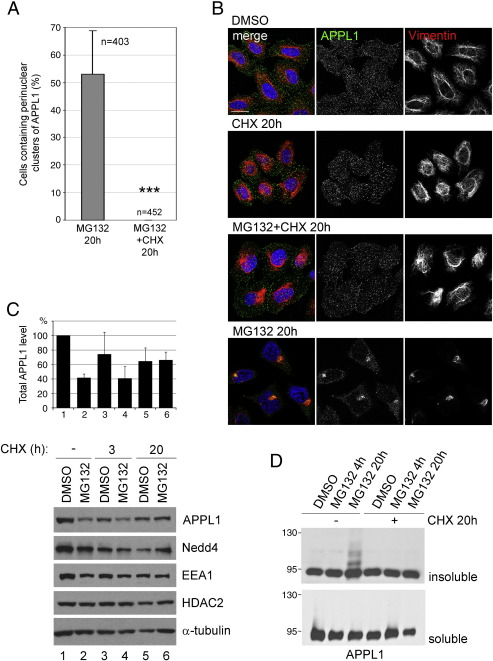
Fig. 7.
Clustering of APPL1 upon MG132 administration depends on protein synthesis. (A) Quantification of the perinuclear clusters of APPL1. HeLa cells were seeded on coverslips and treated with 5 μM MG132 for 20 h with or without 10 μg/ml cycloheximide (CHX). Six random images taken with a maximally opened confocal pinhole were analyzed for each condition; the cells containing clustered APPL1 were counted and expressed as a percentage of all scored cells (n indicates the number of scored cells). The graph shows an average of two independent experiments with standard deviation. ***, p < 0.001 compared with control MG132 alone (Mann–Whitney test). (B) HeLa cells were treated for 20 h with control DMSO, 10 μg/ml cycloheximide, 10 μM MG132 alone or both together. The cells were fixed and stained for APPL1, vimentin and DAPI (blue). Bar, 20 μm. (C) HeLa cells were treated with DMSO or 10 μM MG132 with or without 10 μg/ml cycloheximide for the indicated times prior to lysis in hot sample buffer. Equal protein amounts were resolved on SDS-PAGE followed by detection of levels of the indicated proteins by Western blot. The graph shows quantification of APPL1 total protein levels from Western blot bands, with average values and standard deviation obtained from two separate experiments. Calculation has been performed using ImageJ software. (D) HeLa cells were treated as indicated, lysed in RIPA buffer, centrifuged and extracted into the soluble and insoluble fractions that were subjected to Western blotting using anti-APPL1 antibodies.