Figure 4.
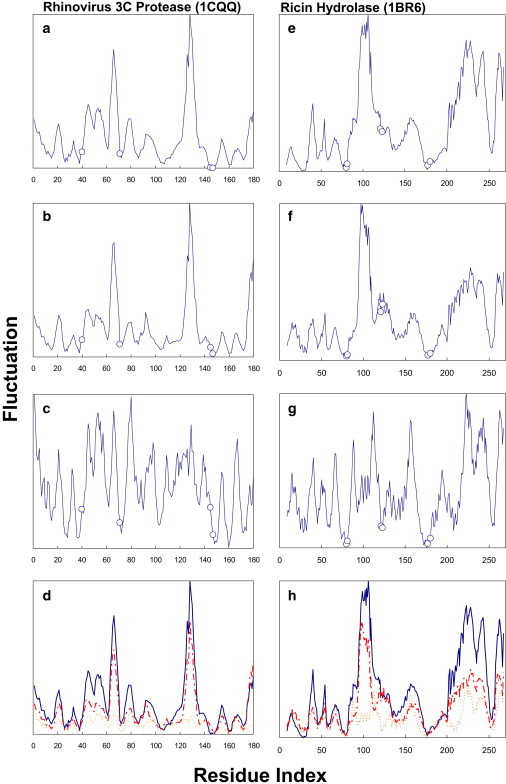
(a–c) The total (a), internal (b), and external (c) fluctuations of the slowest two GNM modes (or the slowest six eGNM modes) are plotted as functions of residue index for rhinovirus 3C protease (1cqq). The amplitudes of the fluctuations are scaled such that the most mobile residue in a given fluctuation profile has the value of unity, which is also the visible maximum in the ordinate of every panel (32). Open circles denote the locations of the active sites H40, D71, G145, and C147. (d) The relative amplitudes of the total (solid line), internal (broken line), and external (dotted line) fluctuations of the slowest two modes are plotted against the residue index. The most mobile residue in total fluctuations, S128, spans the full scale of the ordinate. (e–h) Physical quantities as described in a–d, but for ricin hydrolase (1br6). Active sites Y80, V81, G121, Y123, E177, and R180 are represented by open circles, and their spatial distributions in relation to the rotation axes identified from the first two eGNM modes are shown in Fig. S5.
