Figure 5.
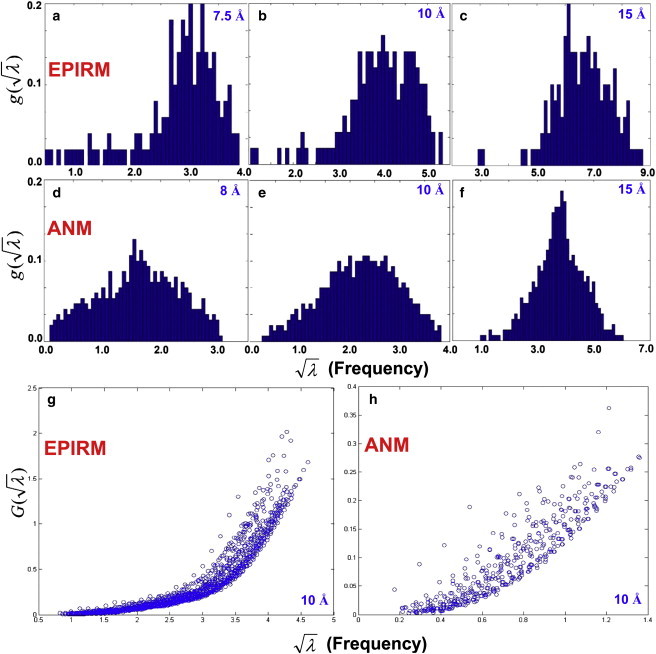
(a–f) Mode density functions g(λ1/2), the number of modes/frequency range divided by the total number of residues in myoglobin (1a6m), are plotted for EPIRM (a–c) and ANM (d–f) using cutoff distances from 7.5 to 15 Å. Here, the number of bins is one-third of the total number of protein residues. (g and h) G(λ1/2), the cumulative mode density functions, are plotted for the slowest 10% of the EPIRM modes (g) and the slowest 4% of the ANM modes (h) at a fixed cutoff distance of 10 Å. Data points are obtained from the 30 high-resolution structures listed in Table S1. All the fitting correlations between G(λ1/2) and λβ/2 for the proteins examined are found to be >0.98.
