Figure 1.
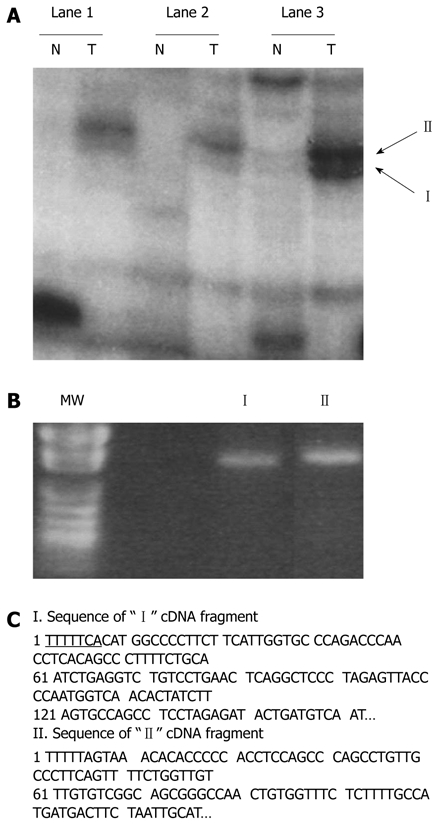
Identification of differentially displayed cDNA fragments from paired hepatocellular carcinoma cells (T) and nontumorous liver tissues (N). A: Differentially displayed PCR products (“I” and “II”) amplified with AP-10 primer and T12MA. Lanes 1-3 denote the three B6C3 F1 mouse samples, respectively; B: Recovered “I” and “II” from the dried DNA sequencing gel reamplified by PCR. MW lane shows the pUC118 DNA fragments cut by HapII (a restriction enzyme) as molecular weight markers; C: Nucleotide sequences of the bands shown in A. Flanking sequences of T12MA primers are underlined. The two insert-containing fragments were sequenced and identified as gene fragments of GLNS and GSTM3 in comparison with those of nucleotide in GenBank.
