Figure 2.
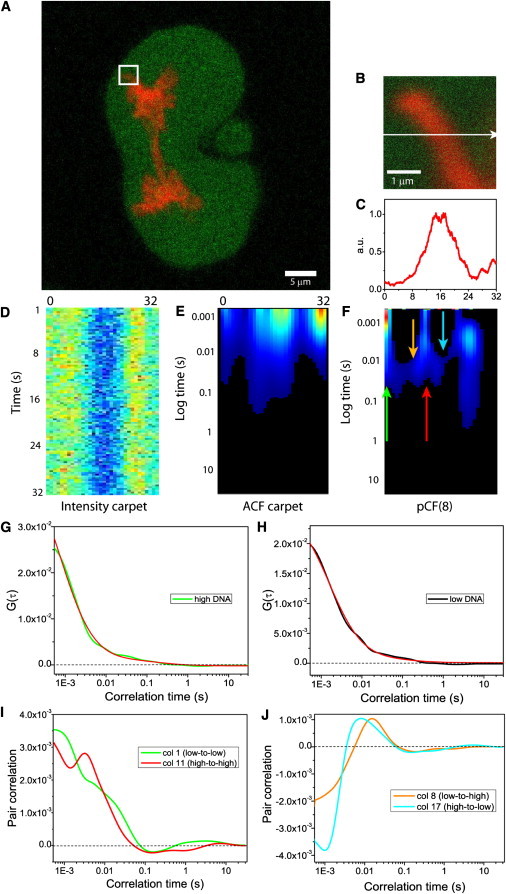
Pair correlation function analysis of intranuclear diffusion of EGFP in a mitotic nucleus. (A) CHO-K1 mitotic cell stably expressing EGFP with the chromosomes marked by H2B-mCherry. (B) Overlay of free EGFP and H2B-mCherry localization in the plane of the 3.3 μm line drawn in the nucleus. (C) Intensity profile of the H2B-mCherry stain across the line measured. (D) Fluorescence intensity carpet of the line drawn across freely diffusing EGFP (200,000 lines). (E) ACF carpet of the line drawn across freely diffusing EGFP (130,000 lines analyzed). (F) The pCF(8) carpet derived for EGFP diffusion between adjacent chromatin density environments across the line measured. The green and red arrows indicate positions where diffusion within the same chromatin density is tested and the orange and cyan arrows indicate positions where diffusion through a change in chromatin density is tested. (G) Fitting of column 15 (which corresponds to high chromatin density) from the ACF carpet in panel E with a two-species model. (H) Fitting of column 30 (which corresponds to low chromatin density) from the ACF carpet in panel E with a one-species model. (I) A plot of the cross correlation function derived from the pCF(8) carpet in panel F for diffusion within a chromatin density environment from the marked arrow positions: low-to-low (column 1 in green) and high-to-high (column 11 in red). (J) A plot of the cross correlation function derived from the pCF(8) carpet in panel F for diffusion through a change in chromatin density from the marked arrow positions: (column 8 in orange) and high-to-low (column 17 in cyan).
