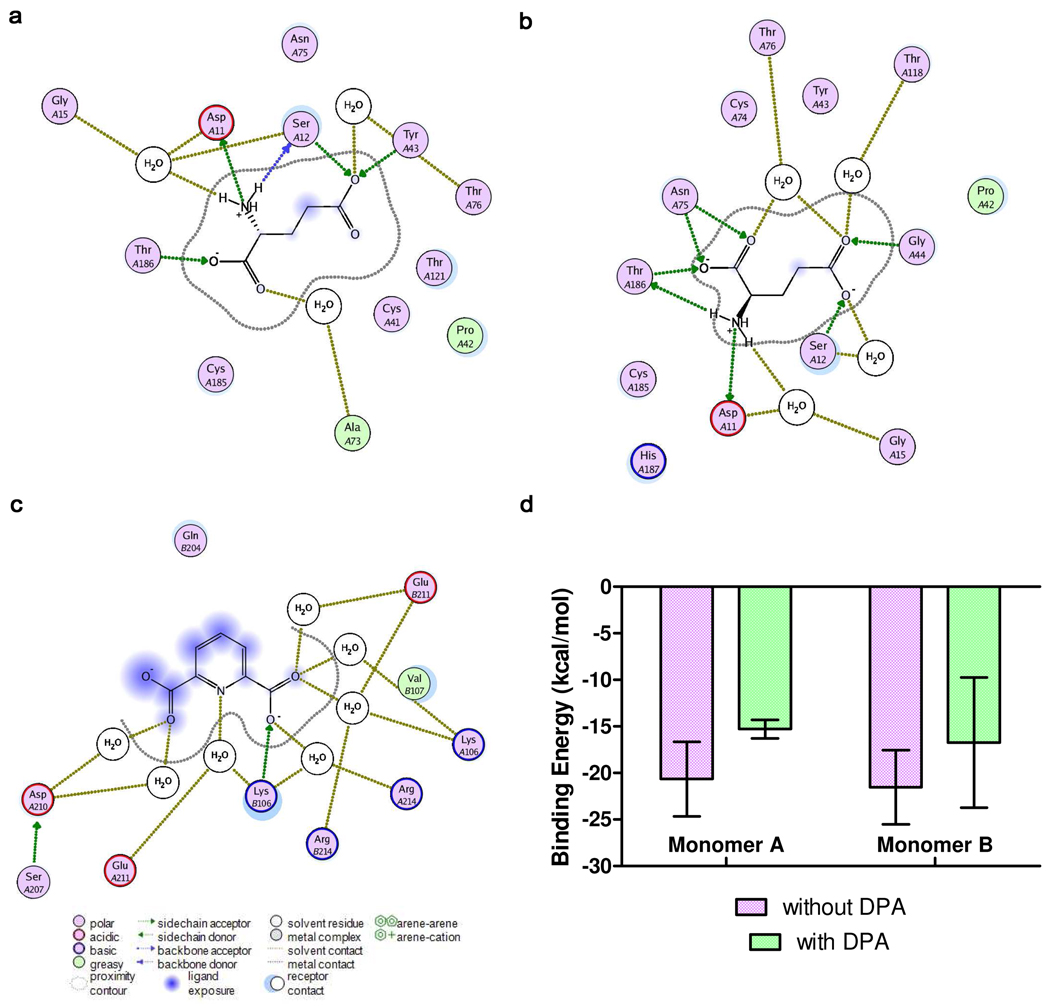
Figure 5.
Ligand interaction maps for glutamate bound to monomer A of the DPA-lacking RacE2 complex (a) and DPA-bound RacE2 complex (b), as well as DPA bound to the cryptic binding site located at the RacE2 dimer interface (c). Maps were generated from the final structures of 20-nanosecond MD simulations using the LigX function of MOE v2009.10. Predicted binding energy of glutamate was averaged over the set of representative structures extracted from MD simulations of the binary (red, n=15) and ternary complex (blue, n=4, d), error bars = SEM. The details of the binding energy calculations are outlined in the Computational Procedures section of Material and Methods.