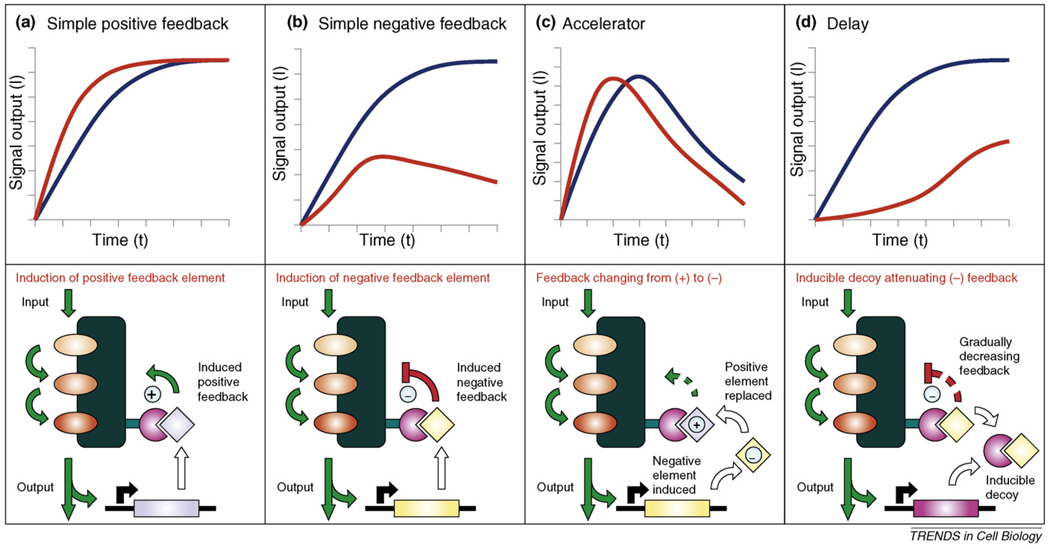
Figure 4.
Scaffold-based network architectures with diverse pathway dynamics. Top panels show experimental kinetic profiles for circuits with wild-type Ste5 (blue) and with a synthetic Ste5 scaffold (red). Lower panels schematically depict underlying circuit architectures. The Ste5 scaffold protein was engineered to include an additional binding site (e.g. by addition of a leuzine zipper, shown in magenta) to enable it to bind other proteins containing a complementary protein interaction element (e.g. another leuzine zipper). By recruiting various effectors or decoys to this scaffolded MAP-kinase module it was possible to change yeast mating pathway kinetics at will [43]. (The original components of this MAPK module: Ste11, Ste7 and Fus3 kinases are coloured in different shades of orange.) (a) Expression of a positive activator (shown in light blue) through a promoter that is responsive to pathway flux creates a simple positive feedback loop, providing more rapid responses compared with wild-type cells. (b) If pathway flux triggers the expression of a negative regulator (yellow), then a simple negative-feedback loop will result. This creates a pulse-like response upon stimulation, unlike wild-type yeast that exhibits a continuous, saturated response. (c) With a careful combination of positive- and negative-feedback elements almost any desired dynamic profile can be realized. A good example is the combination of a constitutively expressed positive regulatory element with a signal-inducible negative element. The two proteins naturally compete with each other for the same binding site, and the inhibitor element will gradually displace the enhancer element as the signalling pathway becomes activated by sustained stimulus. As expected, the system provides a pulse-like output in time, but the peak is shifted towards earlier time points when compared with a simple negative-feedback loop (containing only the inducible inhibitor). Thus, the system behaves as an accelerator. (d) A more complex way to change signalling characteristics can be achieved through the use of decoys – proteins only consisting of binding sites – that, for example, can disrupt association of a constitutively expressed negative regulatory factor with the scaffold. When compared with wild-type cells, a pronounced delay of signal propagation will become apparent. The decoy gradually diminishes the inhibition of signalling as the pathway becomes activated by a sustained stimulus.