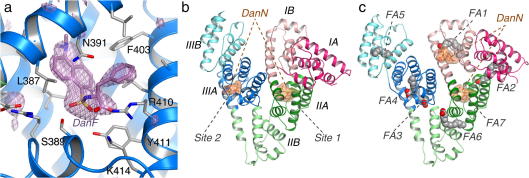
Fig.1.
Location of binding sites for dansylated amino acids determined by X-ray crystallography. (a) Simulated annealing Fo − Fc omit map (contoured at 3σ) showing DanF bound to drug site 2 in sub-domain IIIA. DanF is shown in a stick representation with atoms coloured by atom-type: C – purple; O – red; and N – blue. Sub-domain IIIA is shown in a ribbon representation (blue). Selected protein side-chains are shown as sticks, with carbon atoms coloured grey. (b) Overview of DanN bound to drug sites 1 and 2 in HSA. The ligands are shown in a stick representation with a semi-transparent molecular surface. The protein secondary structure is shown coloured by sub-domain. This colour scheme is maintained throughout. (c) Overview of DanN bound to sub-domains IIA and IB in HSA–myristate. The seven fatty acid binding sites on the protein are labelled FA1–FA7. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)