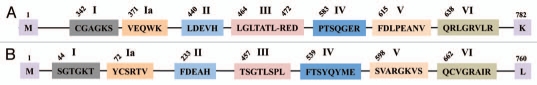
Figure 6.
Schematic representation of seven conserved helicase motifs found in DNA repair helicases, XPB and XPD. The numbers given on top of the diagram indicate amino acid position of the first residue of a conserved motif for the entire protein sequence. (A) Amino acid consensus for motifs of XPB helicase. The sequences representing CGAGKS (motif I), VEQWK (motif Ia), LDEVH (motif II), LGLTATLxRED (motif III), PTSQGER (motif IV), FDLPEANV (motif V) and QRLGRVLR (motif VI) are shown. The first and last protein residues are methionine (M) and lysine (K) respectively are indicated. (B) Amino acid consensus for motifs of XPD helicase. The consensus sequence for these motifs is shown, SGTGKT (motif I), YCSRTV (motif Ia), FDEAH (motif II), TSGTLSPL (motif III), FTSYQYME (motif IV), SVARGKVS (motif V) and QCVGRAIR (motif VI). The first and last protein residues are methionine (M) and leucine (L) are shown.