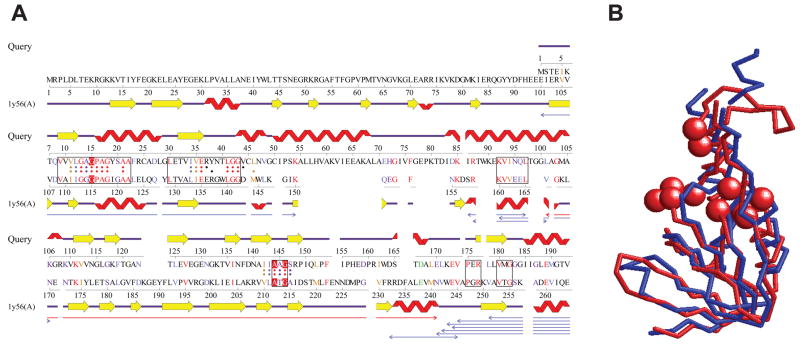
Figure 6.
Structure similarity between the N-terminal domain of E3 and L-proline dehydrogenase α subunit. (A) Structural alignment between the N-terminal domain of the model of E3 (Query) and L-proline dehydrogenase α subunit (PDB CODE: 1y56A) computed by the ProFunc server.30 The homology model of E3 was built with ModBase,29 based on a template structure (PDB CODE: 1lvl). The boxes on the sequences of the two proteins represent segments where the sequence identity exceeds 35%. Dots assigned to each sequence indicate residues within 10 Å to the center of the template protein (1y56A) and hence considered in computing the alignment. The three residues in red comprise of the ATP-binding site of the template, which are initially matched in the ProFunc search. The thin arrows below alignments are structurally similar regions between the two proteins, which can be superimposed within a root mean square deviation (RMSD) of 3.0 Å. The red arrow shows the longest such segment. For more details, see the ProFunc website (http://www.ebi.ac.uk/thornton-srv/databases/profunc/). (B) Structural superimposition of the N-terminal domain of the model of E3 (blue, residue 1–156) and the homologous region of L-proline dehydrogenase (red, residue 100–213). Spheres indicate ATP-binding residues of L-proline dehydrogenase. The RMSD of the two structures is 3.48Å.