Abstract
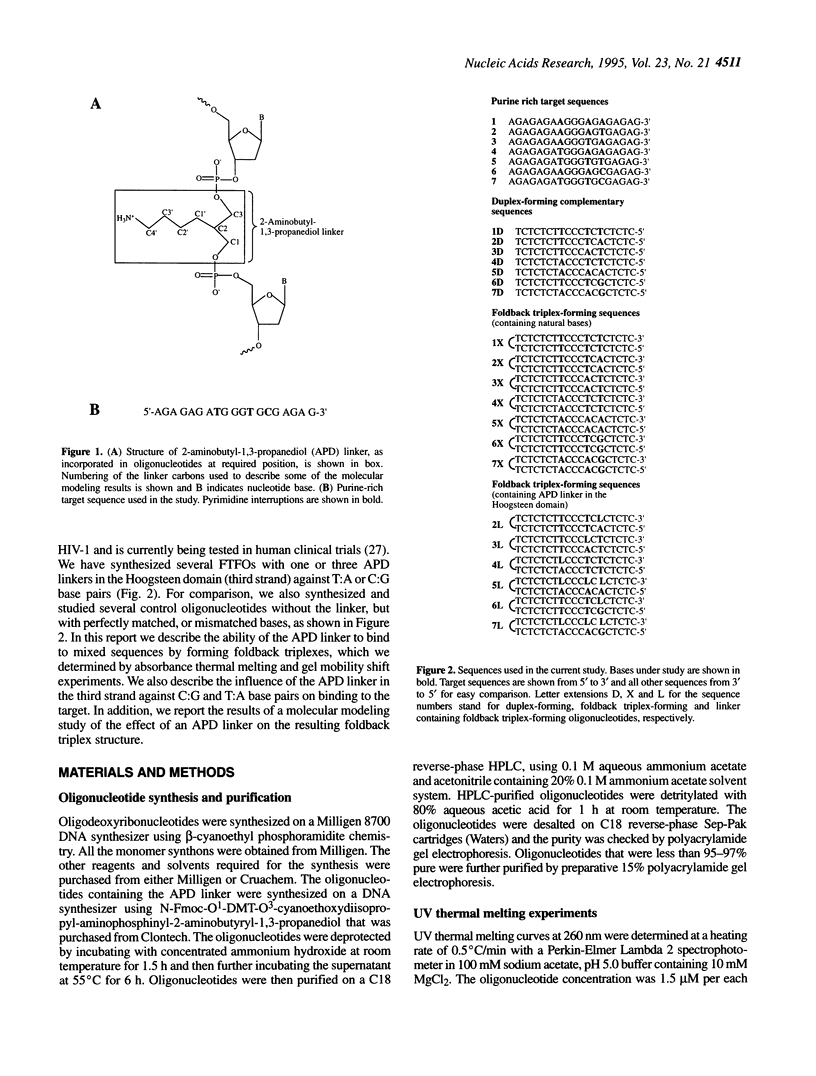
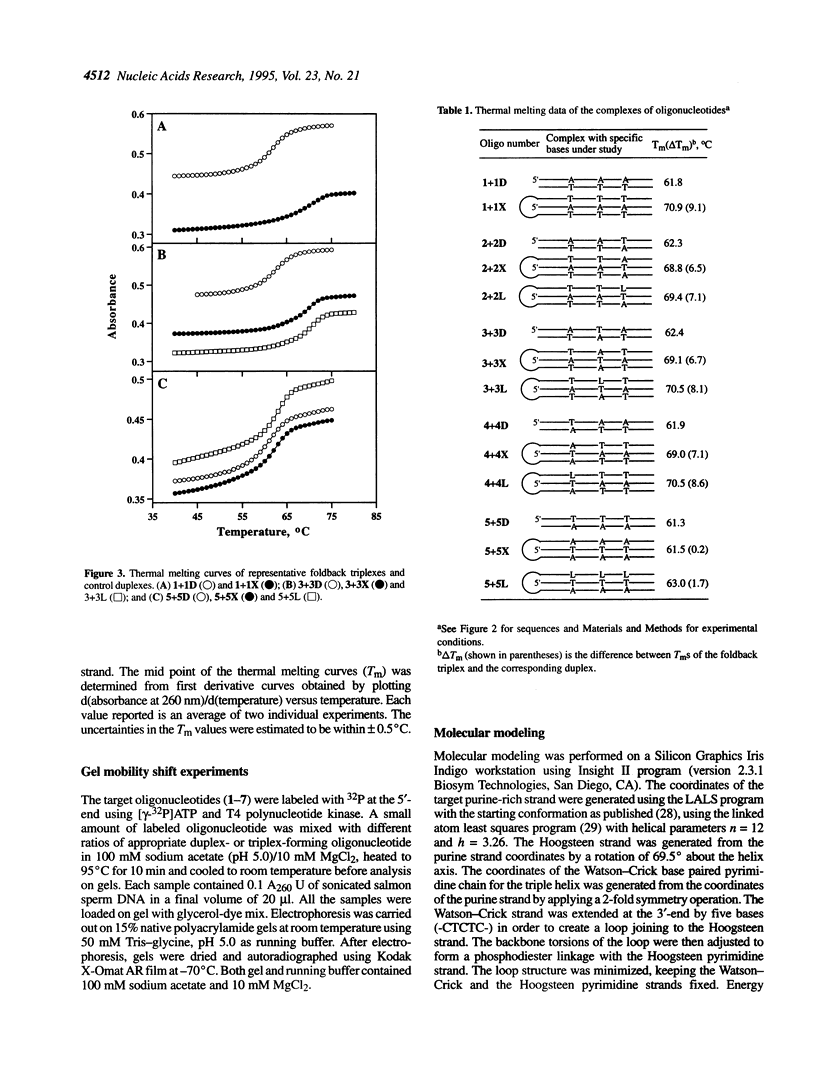
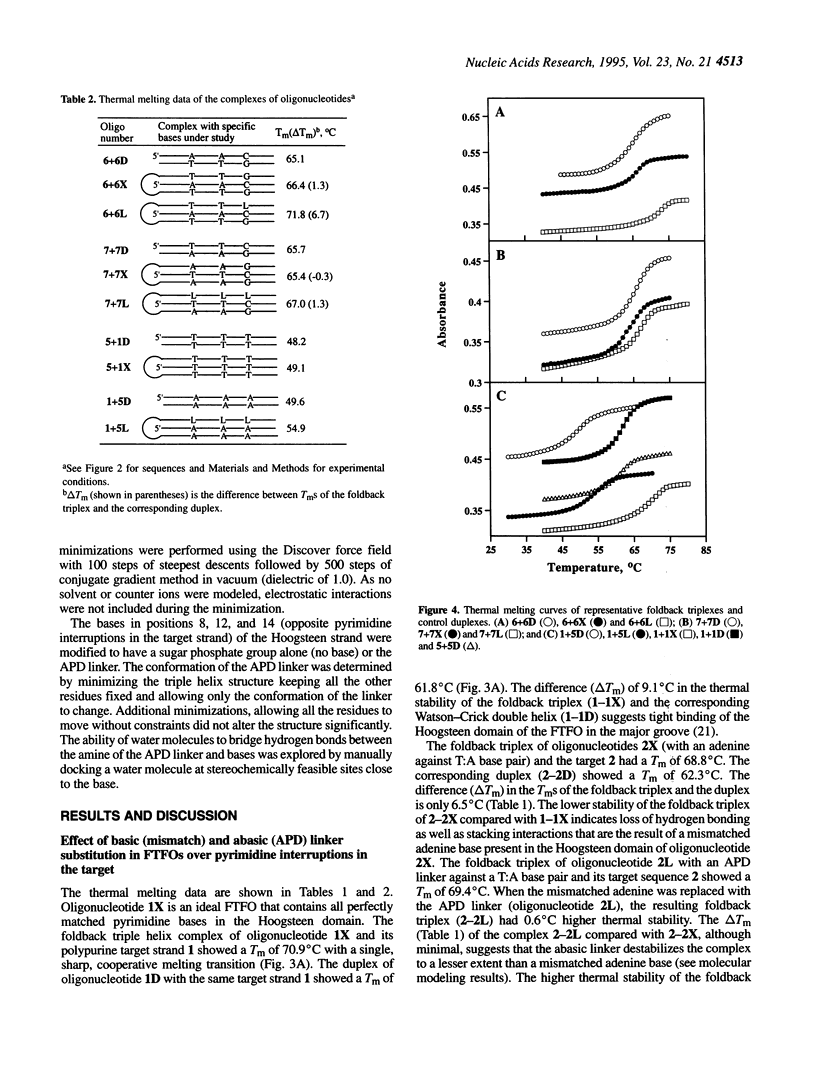
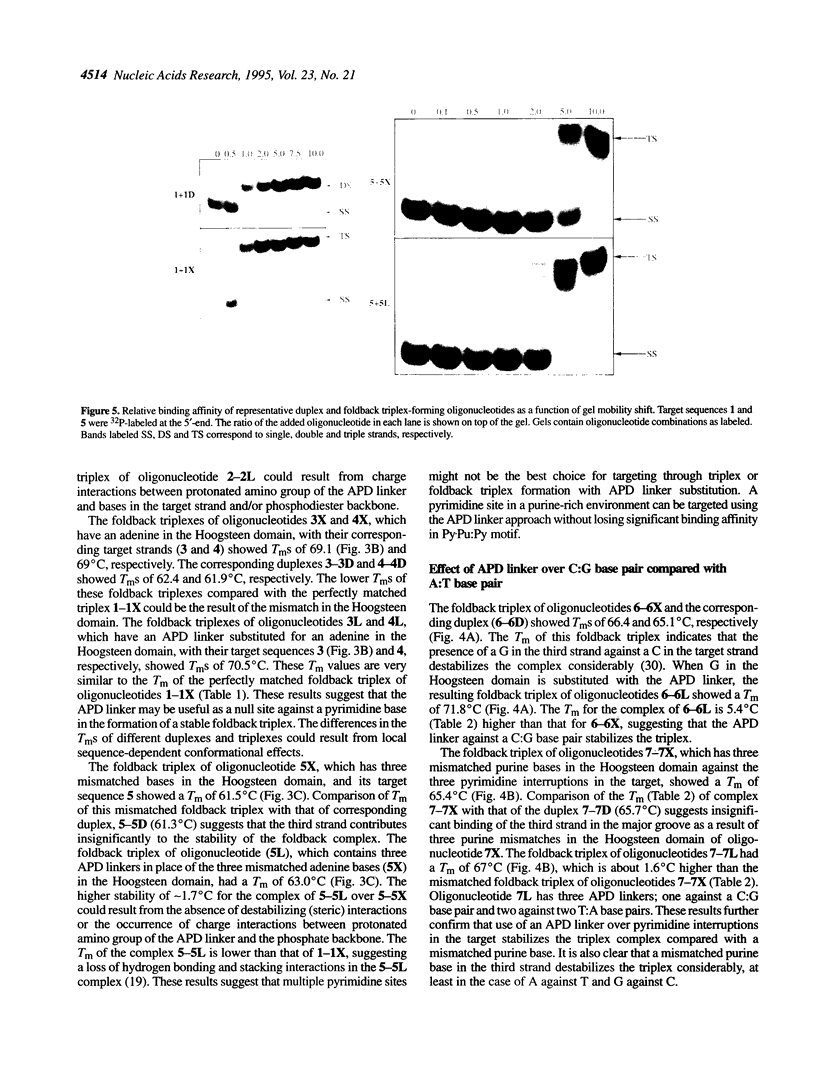
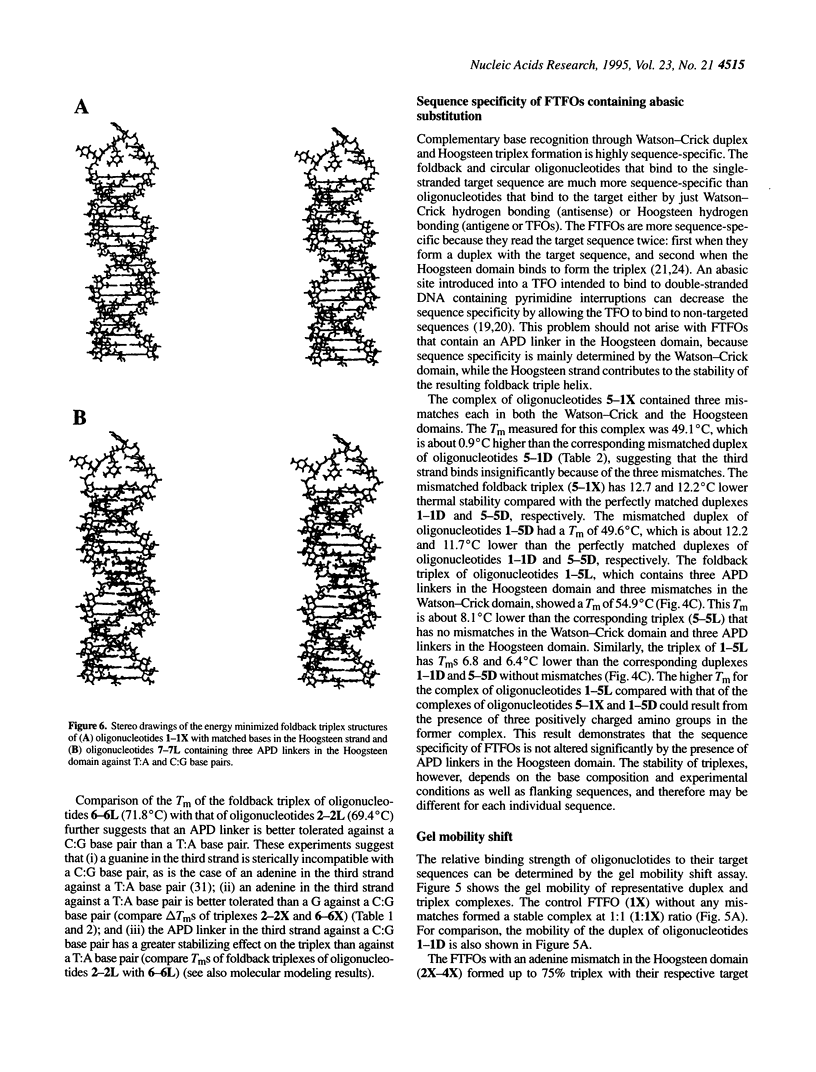
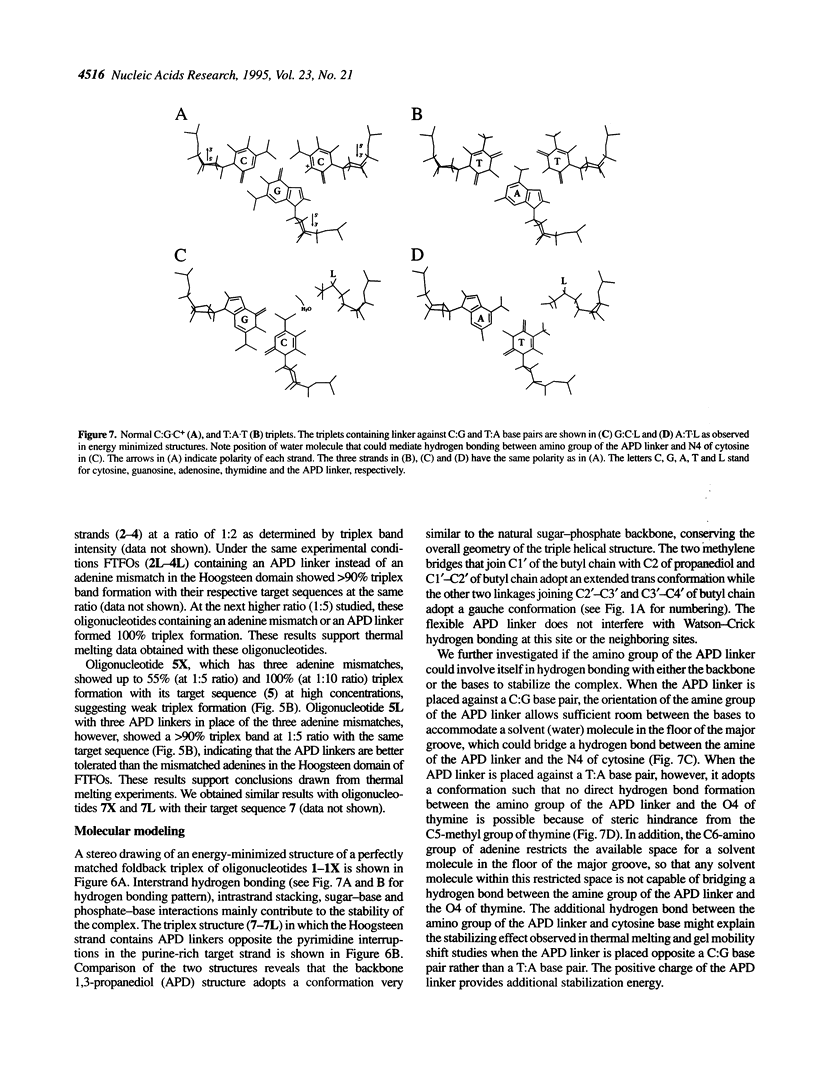
Foldback triplex-forming oligonucleotides (FTFOs) that contain an abasic linker, [2-(4-aminobutyr-1-yl)-1,3-propanediol] (APD linker), in the Hoogsteen domain against pyrimidine bases of a C:G and a T:A base pair were studied for their relative stability and sequence specificity of triplex formation. In general, the APD linker has less destabilizing effect against a C:G base pair than a T:A base pair. Incorporation of three APD linker moieties resulted in decreased binding to the target, which was comparable to results observed with three imperfectly matched natural base triplets. The APD linker incorporation did not result in the loss of sequence specificity of FTFOs, unlike in the case of normal triplex-forming oligonucleotides (TFOs). The introduction of a positively charged abasic linker, however, resulted in decreased stability of the triplex, because of loss of hydrogen bonding and stacking interactions in the major groove. The results of a molecular modeling study show that APD linker can be readily incorporated without any change in the conformation of the natural sugar-phosphate backbone conserving overall triple helix geometry. Further, the modeling study suggests a hydrogen bond formation between the amino group of linker and N4 of cytosine mediated by a solvent molecule (water) in the floor of the base triplet in addition to a contribution from the positive charge on the APD linker amino group. Either a direct or water-mediated hydrogen bond between the amino group of the APD linker and the O4 of thymine is unlikely when the linker is placed against a T:A base pair.
Full text
PDF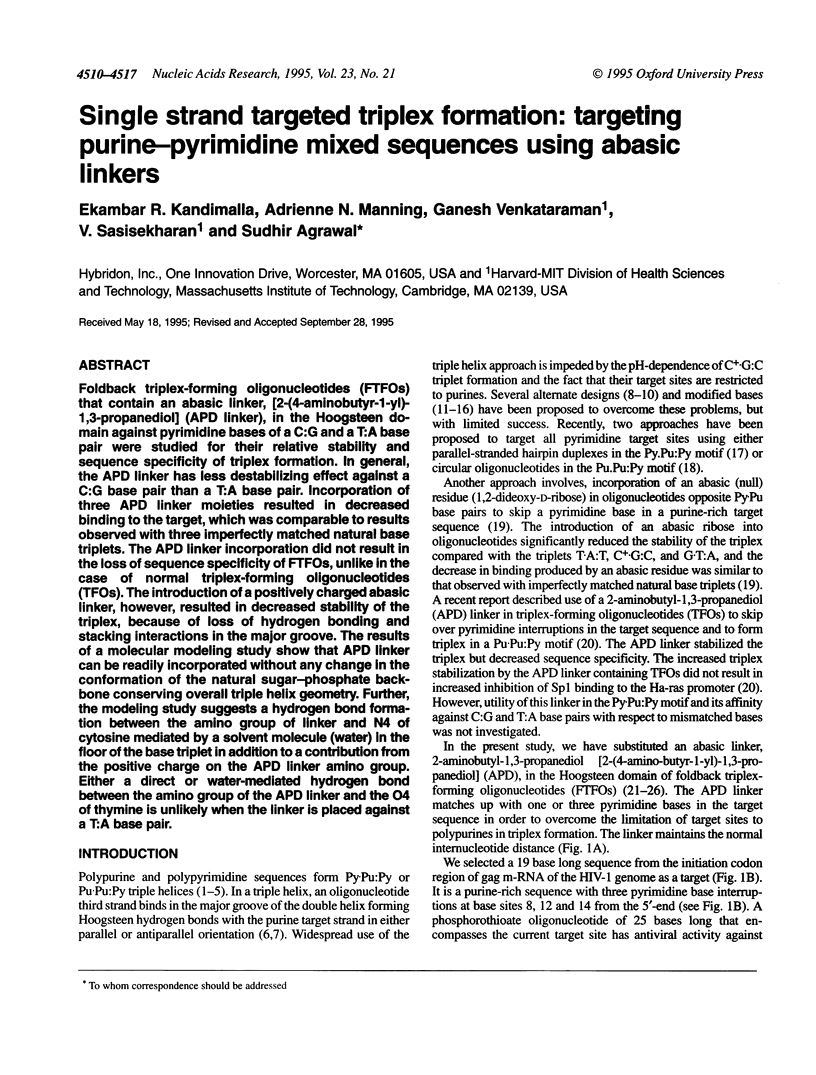

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Agrawal S., Tang J. Y. GEM 91--an antisense oligonucleotide phosphorothioate as a therapeutic agent for AIDS. Antisense Res Dev. 1992 Winter;2(4):261–266. doi: 10.1089/ard.1992.2.261. [DOI] [PubMed] [Google Scholar]
- Cheng Y. K., Pettitt B. M. Stabilities of double- and triple-strand helical nucleic acids. Prog Biophys Mol Biol. 1992;58(3):225–257. doi: 10.1016/0079-6107(92)90007-s. [DOI] [PubMed] [Google Scholar]
- Cooney M., Czernuszewicz G., Postel E. H., Flint S. J., Hogan M. E. Site-specific oligonucleotide binding represses transcription of the human c-myc gene in vitro. Science. 1988 Jul 22;241(4864):456–459. doi: 10.1126/science.3293213. [DOI] [PubMed] [Google Scholar]
- Griffin L. C., Dervan P. B. Recognition of thymine adenine.base pairs by guanine in a pyrimidine triple helix motif. Science. 1989 Sep 1;245(4921):967–971. doi: 10.1126/science.2549639. [DOI] [PubMed] [Google Scholar]
- Horne D. A., Dervan P. B. Effects of an abasic site on triple helix formation characterized by affinity cleaving. Nucleic Acids Res. 1991 Sep 25;19(18):4963–4965. doi: 10.1093/nar/19.18.4963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayasena S. D., Johnston B. H. Sequence limitations of triple helix formation by alternate-strand recognition. Biochemistry. 1993 Mar 23;32(11):2800–2807. doi: 10.1021/bi00062a010. [DOI] [PubMed] [Google Scholar]
- Jetter M. C., Hobbs F. W. 7,8-Dihydro-8-oxoadenine as a replacement for cytosine in the third strand of triple helices. Triplex formation without hypochromicity. Biochemistry. 1993 Apr 6;32(13):3249–3254. doi: 10.1021/bi00064a006. [DOI] [PubMed] [Google Scholar]
- Kandimalla E. R., Agrawal S. Single strand targeted triplex-formation. Destabilization of guanine quadruplex structures by foldback triplex-forming oligonucleotides. Nucleic Acids Res. 1995 Mar 25;23(6):1068–1074. doi: 10.1093/nar/23.6.1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandimalla E. R., Agrawal S. Single-strand-targeted triplex formation: stability, specificity and RNase H activation properties. Gene. 1994 Nov 4;149(1):115–121. doi: 10.1016/0378-1119(94)90419-7. [DOI] [PubMed] [Google Scholar]
- Lyamichev V. I., Mirkin S. M., Frank-Kamenetskii M. D., Cantor C. R. A stable complex between homopyrimidine oligomers and the homologous regions of duplex DNAs. Nucleic Acids Res. 1988 Mar 25;16(5):2165–2178. doi: 10.1093/nar/16.5.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher L. J., 3rd, Wold B., Dervan P. B. Inhibition of DNA binding proteins by oligonucleotide-directed triple helix formation. Science. 1989 Aug 18;245(4919):725–730. doi: 10.1126/science.2549631. [DOI] [PubMed] [Google Scholar]
- Mayfield C., Miller D. Effect of abasic linker substitution on triplex formation, Sp1 binding, and specificity in an oligonucleotide targeted to the human Ha-ras promoter. Nucleic Acids Res. 1994 May 25;22(10):1909–1916. doi: 10.1093/nar/22.10.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mergny J. L., Sun J. S., Rougée M., Montenay-Garestier T., Barcelo F., Chomilier J., Hélène C. Sequence specificity in triple-helix formation: experimental and theoretical studies of the effect of mismatches on triplex stability. Biochemistry. 1991 Oct 8;30(40):9791–9798. doi: 10.1021/bi00104a031. [DOI] [PubMed] [Google Scholar]
- Miller P. S., Bhan P., Cushman C. D., Trapane T. L. Recognition of a guanine-cytosine base pair by 8-oxoadenine. Biochemistry. 1992 Jul 28;31(29):6788–6793. doi: 10.1021/bi00144a020. [DOI] [PubMed] [Google Scholar]
- Moser H. E., Dervan P. B. Sequence-specific cleavage of double helical DNA by triple helix formation. Science. 1987 Oct 30;238(4827):645–650. doi: 10.1126/science.3118463. [DOI] [PubMed] [Google Scholar]
- Ono A., Chen C. N., Kan L. S. DNA triplex formation of oligonucleotide analogues consisting of linker groups and octamer segments that have opposite sugar-phosphate backbone polarities. Biochemistry. 1991 Oct 15;30(41):9914–9912. doi: 10.1021/bi00105a015. [DOI] [PubMed] [Google Scholar]
- Praseuth D., Perrouault L., Le Doan T., Chassignol M., Thuong N., Hélène C. Sequence-specific binding and photocrosslinking of alpha and beta oligodeoxynucleotides to the major groove of DNA via triple-helix formation. Proc Natl Acad Sci U S A. 1988 Mar;85(5):1349–1353. doi: 10.1073/pnas.85.5.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghunathan G., Miles H. T., Sasisekharan V. Symmetry and molecular structure of a DNA triple helix: d(T)n.d(A)n.d(T)n. Biochemistry. 1993 Jan 19;32(2):455–462. doi: 10.1021/bi00053a009. [DOI] [PubMed] [Google Scholar]
- Xodo L. E., Manzini G., Quadrifoglio F. Spectroscopic and calorimetric investigation on the DNA triplex formed by d(CTCTTCTTTCTTTTCTTTCTTCTC) and d(GAGAAGAAAGA) at acidic pH. Nucleic Acids Res. 1990 Jun 25;18(12):3557–3564. doi: 10.1093/nar/18.12.3557. [DOI] [PMC free article] [PubMed] [Google Scholar]



