Figure 1.
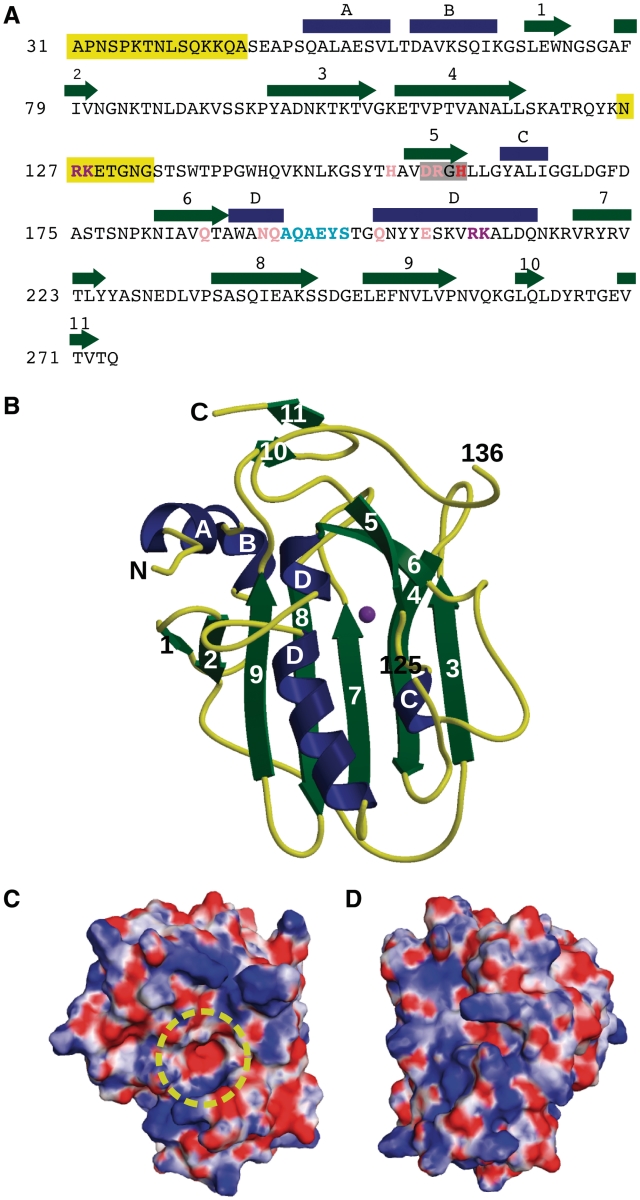
Sequence and structure of EndA. (A) Sequence and secondary structural elements of EndA from S. pneumoniae. α-Helices are indicated as blue rectangles and β-strands as green arrows. The DRGH motif is boxed in gray. The position of the general base (His160) is shown in red. Mutated residues involved in catalysis are shown in pink. Residues likely involved in DNA substrate binding and/or catalysis are shown in purple. The ‘finger-loop’ is shown in cyan. Disordered regions are highlighted in yellow. (B) Ribbon diagram of EndA, showing the ‘front face’ of the molecule that contains the active site. Secondary structural elements are color-coded as in (A) and the magnesium ion is colored purple. The positions of the termini and the ends of the disordered loop are labeled. (C) Electrostatic surface potential of the ‘front face’ of EndA, in the same orientation as in (B). The surface was calculated using the Adaptive Poisson–Boltzmann Solver tool in PyMOL (47). The potential ranges from −8 kTe−1 (red) to 8 kTe−1 (blue). The location of the active site is highlighted by a yellow circle. (D) Electrostatic surface potential of the ‘back face’ of EndA [Disordered side chains lacking density were modeled onto the EndA surface for panels (C) and (D).].