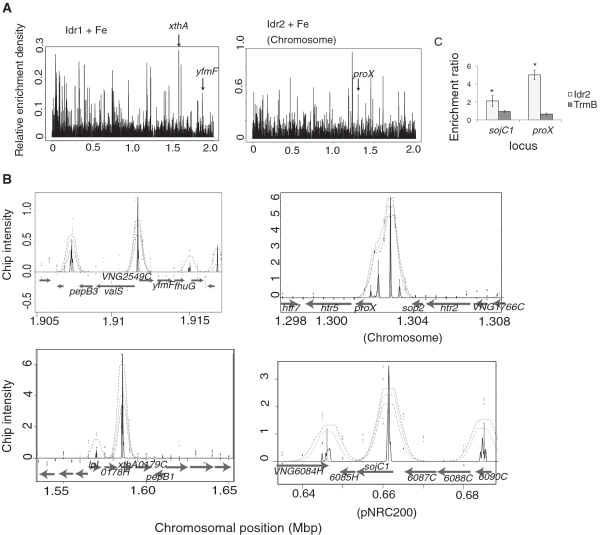
Figure 3.
Idr1 and Idr2 bind preferentially in the presence of iron to loci throughout the genome. (A) ChIP–chip binding peaks at sites across the genome. (Left) All Idr1 binding locations detected in the five biological replicate ChIP–chip experiments are depicted as black peaks. (Right) ChIP–chip data are shown for Idr2. On the y-axis, relative enrichment represents the scaled peak intensity for all replicates combined with a density algorithm (‘Materials and Methods’ section). The position of each peak on the chromosome is shown on the x-axis in megabase pairs (Mbp). (B) Representative examples of peaks from the MeDiChI algorithm output. Idr1 targets (top, siderophore transport operon; bottom, xthA endonuclease) are shown on the left. Idr2 targets are shown on the right (top, proX transport operon in the chromosome; bottom, sojC1 cell division protein on the pNRC200 plasmid). Gray arrows depict the direction and location of ORFs along the segments of the chromosome shown on the x-axis. Each ORF is labeled according to its function. Some of these functions are discussed in the text; others are listed in Supplementary Table S2. Gray dotted lines in each peak show the bootstrap confidence range from the MeDiChI model fit, with the innermost line representing the highest confidence. Black peaks represent the best fit to the model (36). Black dots represent raw enrichment ratio data points from representative ChIP–chip experiments. (C) ChIP–qPCR validation of Idr2 binding sites. Primers surrounding the most intense black peaks shown in (B) were used in quantitative PCR experiments to validate the observed ChIP-chip peaks. Error bars represent the ± SEM from the mean of 12 replicate trials. Asterisks represent significant differences (P ≤ 0.05) between the binding enrichments for TrmB compared to those for Idr2.